On high-throughput sample processing and laboratory techniques: concerns, pitfalls and more

Centuries of scientific research and development have led to techniques that have set the foundation for the future of automated research with high-throughput sample processing (HTSP). The world is facing challenges from climate change to antibiotic resistance that require collaborative research to find answers and potential solutions. These large-scale collaborations can generate vast sample numbers, into the thousands or tens of thousands, which require substantial infrastructural innovation to enable efficient and reliable processing. With evidence now mounting that longitudinal time-series studies are needed to understand microbiome dynamics, and to capture relevant microbial biomarkers of relevance to human or environmental health, the number of samples required to test next-generation hypotheses will only increase.
There are many known solutions to improve the throughput and cost of sample processing. HTSP requires the automation of experimental components to ensure that large-scale studies become feasible and reproducible, while requiring less human effort and less resources (e.g., consumables and reagents) to complete. As a result, HTSP also accelerates the development of standard protocols, which are essential for reproducibility and can aid democratization of technologies and techniques, providing the potential to improve the reliability and quality of data. Good examples of this include the Human Microbiome Project and the Earth Microbiome Project, whose standard protocols have shaped an entire field. Standardization also facilitates increases in data quality by enabling more robust statistical checks on the larger datasets. To enable these advantages, it is essential that we continue to develop high-throughput approaches, which means overcoming hurdles, developing detailed records of processing steps, creating new collaborations especially across disciplines, and embracing modularity to improve the flexibility of processing pipelines to accept new technologies.
HTSP has revolutionized sequencing, genotyping, cell biology, screening, and imaging. The most visible success story is high-throughput sequencing technologies, which have revolutionized microbiome studies, enabling statistical associations between microbial phenotypes and disease states that were previously invisible. Amplicon sequencing was enabled at scale purely through high-throughput sequencing, providing an extremely low cost per sample for analysis. Indeed, the development of highly curated platforms with robotic automation for generating high-quality sequencing libraries has enabled rapid turn-around, and improved standardization and reagent use efficiencies.
HTSP in any laboratory requires a delicate balance between speed, volume and quality. Many recent advances have substantially increased the speed and quantity of samples that can be processed, but to ensure quality requires advanced skills within the research lab. Akin to having ‘green fingers’ many of the new robots and pipelines require expert handling. This leads to significant inter-lab variability, reducing the beneficial impact of standardization. The differences in sample processing quality are most obvious between research labs and service contract labs, primarily due to the quality and quantity of the experience of the team. For example, if a student researcher needs to perform a task for which they have little experience and lack the appropriate equipment, it can lead to spurious results that introduce substantial variability in data output across studies. Service contract labs with extensive technical staff typically already have a validated protocol ready and automation to handle samples in a more controlled environment, thus improving the success and standardization of output. The key to success is consistency across all protocols, and the use of standards, positive controls and quality checks during the process to account for variance within and between large studies, which academic laboratories often lack. Therefore, collaboration with an experienced research group or a service contract facility is necessary in order to avoid pipeline biases and increase the quality of sample processing and data generation.
- RNA Sequencing; as accurate as once believed?
- The rise of metascience
- Fake news, the reproducibility crisis & politics – science communication with Gareth Mitchell
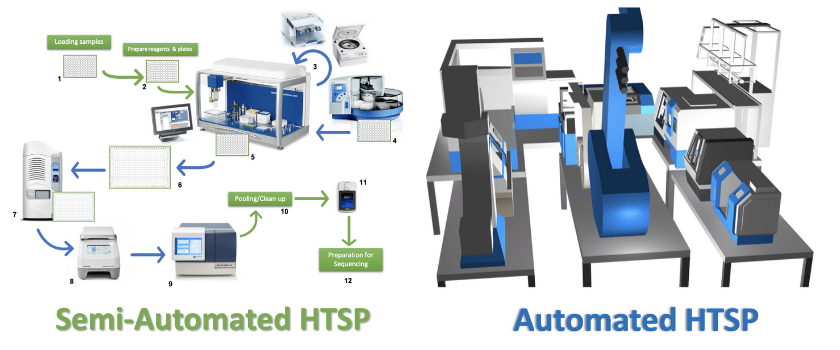
In a semi-automated HTSP lab, samples are no longer manually transferred between 96 and 384 well plates, but instead are processed using liquid handling robotics, which can also handle PCR, amplicon quantification, and DNA concentration for library preparation and sequencing (Figure 1). Academic labs often employ both manual and automated steps due to the cost of the equipment, as well as providing the flexibility to change steps when a pipeline has not been fully optimized for one process. Also, academic labs may not always need HTSP; when dealing with sample sizes of <1000 it is often easier to just manually handle many of the steps. The green arrows in Figure 1 represent manual processing steps, such as loading samples into a 96-well plate, preparing reagents and plates, and transferring the plates between equipment. Loading and transferring sample plates manually increases the level of sample handling, which increases the risk of contamination due to human error. In Figure 1, the blue arrows and equipment already include automated steps. While pharma and biotech industries have already integrated fully automated pipelines into chemical and drug manufacturing, the future of molecular biology and microbiome research would benefit significantly from the reproducibility, efficiency and quality provided by full automation. It would reduce the tedium associated with massive sample processing, as well as mitigating mistakes, allowing for quality control check points, and it would substantially speedup data delivery for collaborators. For instance, in the fully automated microbiome core, only the robot would interact with the processing equipment, reducing the risk of contamination, allowing streamlining of each protocol. This will allow the technician to oversee and optimize the process, rather than handling the samples throughout, significantly increasing the likelihood of consistent and reproducible results. The use of automation with specialized techniques and miniaturized assays drives down the cost and the time required for processing, allowing scientists to address biological questions that are otherwise unattainable using conventional methods.
Despite the power of HTSP technology platforms to improve standardization, minimize mistakes and streamline costs, the main pitfall remains cross-contamination between samples. Whether you are running a protocol with single tubes, 96-well plates, or beyond, it is critical to have adequate labelling, proper controls and to avoid contact between samples. However, even with the most stringent preventative measures in place, exposure to the ubiquity of environmental microbes is probably unavoidable. Therefore, it is virtually impossible to maintain a completely sterile space for sample preparation, especially when the samples you are processing are a major source of potential contaminants. It is essential that the community accepts standards intended to better monitor and quantify these inevitable sources of noise in any given study, from the sample repository to the sequencing platform. In addition to external and cross contamination, technical hurdles such as DNA extraction efficiency and molecular probe biases create noise that is also essentially unavoidable and as such needs to be appropriately accounted for.
However, increased automation creates specialized roles in the laboratory, so that a laboratory technician with a broad molecular skill set now must oversee primarily the maintenance of complex robot platforms to ensure that the computational script and mechanical operation runs as expected. This has the potential to lead to a generation of technicians who understand the robots, but not the process the robots are mediating. When the robots breakdown, lab activities can grind to a halt, as the system is now dependent on these operations. Small labs cannot afford to hire specialized people, nor to cover the cost of operational redundancies to avoid such downtime. Expensive robot platforms require constant maintenance and routine engineering. When the system breaks down in the middle of an operation, it often means that those samples are lost, and the whole system needs to be re-run.
A possible solution to ensure a reduction in systems failure and costly operation downtime could lie in the use of machine learning techniques. A standard robotic platform outputs megabytes of data on a single operation. Harnessing these data to predict when part of the equipment platform will fail would provide an opportunity to revolutionize access to these systems by reducing the dependency on a redundant system. Machine learning and artificial intelligence systems are helpful for navigating the decision-making process involved in cost vs benefit comparison in research outcomes. While it seems as if the benefits outweigh the drawbacks, it is crucial that the development of new automation incorporates the user to reduce the likelihood that we create an army of black boxes that could harm future innovation in pipelines. New protocols must also include appropriate controls, quality checks and stop-go decision points to avoid errors throughout the pipeline. We must embrace integration of computer scientists into the primary lab process to ensure that machine-readable data are produced and harnessed effectively to improve our ability to predict failures and refinements in the system. The hurdles experienced in laboratory techniques and during massive sample processing still indicate the importance of existing research skills, in addition to the new skills that the surge of revolutionary technologies will require in the future.