Sylvia Boj on gene editing in organoids and their use in drug discovery
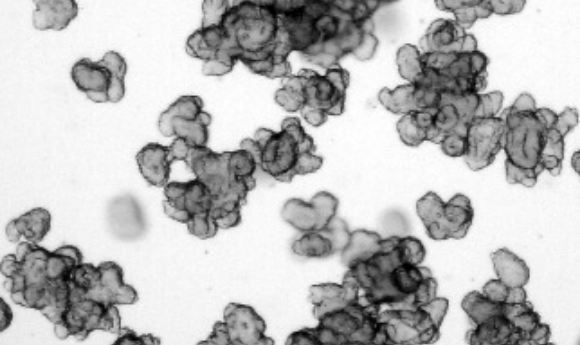
In this interview we catch up with Sylvia Boj, who takes us through a recent paper that used patient-derived organoids as a test case for a new CRISPR technique and talks about the impact this technology could have in the development of organoids for regenerative medicine. We also discuss future developments in organoids, and their involvement in drug discovery and cancer research.
 Sylvia Boj is the Chief Research Officer at Hubrecht Organoid Technology (HUB; Utrecht, The Netherlands). Her primary responsibility is to lead the HUB research team and to coordinate all the research activities that are taking place in the organization.
Sylvia Boj is the Chief Research Officer at Hubrecht Organoid Technology (HUB; Utrecht, The Netherlands). Her primary responsibility is to lead the HUB research team and to coordinate all the research activities that are taking place in the organization.
Can you tell us about the updated gene editing technique developed from CRISPR technology that you recently applied in cystic fibrosis organoid models?
In this publication our academic collaborators from the Beekman group at University Medical Center Utrecht and the Clevers group at the Hubrecht Institute (both Utrecht, The Netherlands ), who are also the main authors of the manuscript, applied a recently developed Cas9 fusion protein. This new generation of Cas9 fusion proteins can introduce changes into genomic DNA, while circumventing some of the side effects that have been observed by initially developed CRISPR–Cas9 proteins, which were more prone to introduce some deleterious off-target double strand breaks.
The idea was to evaluate the activity of two new Cas9 base editors (SP Cas9 and XSP Cas9) in a very clinically relevant model for cystic fibrosis, intestinal organoids derived from cystic fibrosis patients, with a very easy readout, the forskolin induced swelling (FIS) assay, in which restoration of CFTR function can be easily measured. So, the cystic fibrosis organoid models were used as proof-of-concept for the new CRISPR technique.
Could these new Cas9 proteins help alter aspects of cystic fibrosis or other organoids that you were previously unable to manipulate?
In the context of cystic fibrosis and organoid technology, it is slightly more difficult to reflect on how this new generation of Cas9 base editors can make an impact, but they definitely have an incredible impact in applications for regenerative medicine.
Together with the intestinal model, liver organoids, derived from human biopsies, have shown their potential to engraft and regenerate tissue. We can, therefore, foresee new Cas9-editing based technologies repairing mutations in metabolic genes in liver organoids generated from patients with such metabolic diseases. Repaired organoids could then be transplanted back to the patient to replace the mutant live cells with the repaired ones. This clearly requires a high level of accuracy to ensure existing mutations are fixed without inducing new, off-target mutations in the organoids that are intended to be used for transplant tissue.
What were some of the most exciting discoveries for you from that study?
I think it is very exciting that we have proven in a relevant clinical model that this new generation of Cas9 proteins can repair a gene and do not introduce off-target effects in the genome. The possibility that with this approach could open a window for the genetic-based treatment of certain cystic fibrosis patients. We provided very strong data that pushes forward this approach in terms of improving the treatment of patients with cystic fibrosis, for whom the available drugs cannot restore CFTR function.
What areas you working on in organoid development or organoid application that you find particularly exciting?
The research of organoids in cystic fibrosis is our hallmark and it is the model where we have been able to cover the entire circle in terms of drug development and proving the clinical correlation between organoid models and patient response. This has helped cystic fibrosis organoid models to become an essential in-vitro model for preclinical phases. It is very exciting for us to see that any current drug development in cystic fibrosis is taking place using our organoid models. Furthermore, because of their predictive value, we have been able to include patients in clinical trials where we could identify the optimal treatment for the patients.
We are now working at HUB to translate this success into the field of oncology; this means that we are validating the predictive value of organoids for different types of cancer. We are also using organoids in drug discovery and drug development to find better treatment using a more patient relevant model.
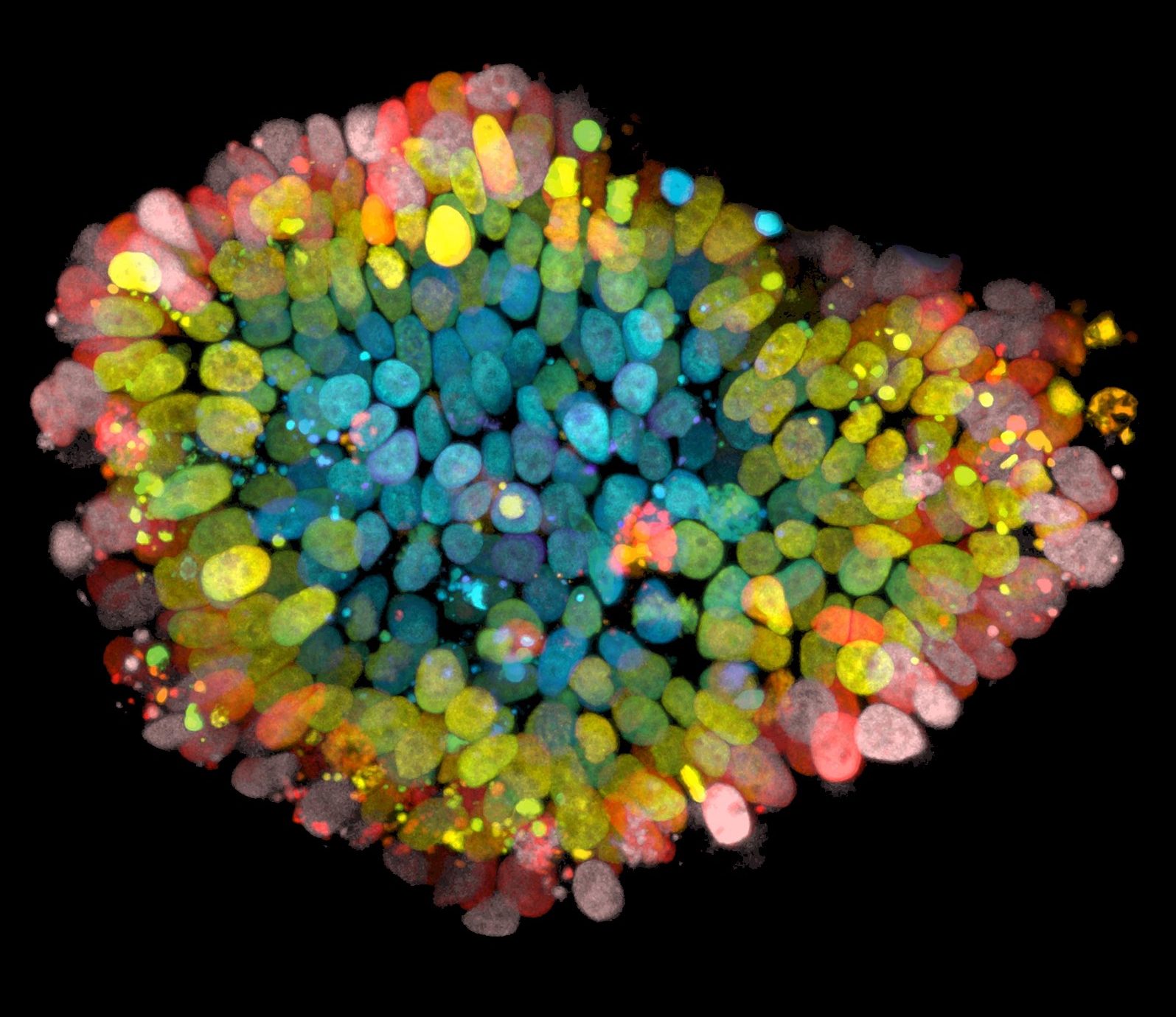 Talking Techniques | Rob Vries on organoids for drug discovery and the study of COVID-19
Talking Techniques | Rob Vries on organoids for drug discovery and the study of COVID-19
BioTechniques Digital Editor, Tristan Free, speaks to Rob Vries, CEO of HUB. We explore the HUB’s use of organoids for drug development and to further study and improve the clinical treatments of diseases such as cystic fibrosis.
What methods are most effective for using organoids for drug discovery? What challenges do you encounter when using organoids for drug discovery?
For drug development, the most usual readout that we perform in drug-screenings is cell viability. We expose the organoids to a certain drug at certain concentrations and then evaluate the impact of the drug on the viability of the organoids.
A particularly challenging aspect that we encountered while using Cas9 technology in cystic fibrosis organoids was the need to target the stem cell – in this case the stem cells of the intestinal organoid from the cystic fibrosis patients – to generate the clone that has repaired the mutation.
The challenge in oncology research is that to make the tumor organoid model truly representative, you need to mimic the heterogeneity of the tumor and this heterogeneity needs to be preserved. When establishing tumor organoids, we preserve multiple different clones to ensure this heterogeneity. However, this Cas9 application requires a single-cell step in order to efficiently introduce Cas9 into the cells. Due to this single-cell step, you lose the heterogeneity from the clones that are generated after it.
What technologies do you employ to carry out that drug screening? Do you have any tips for best practice when using those technologies?
One of our missions is to translate this organoid technology to industry and clinics and to make it as broad as possible, because we believe it’s a very relevant model for drug development. We put a lot of effort into standardizing our protocols and making them very robust. In terms of drug screens, we developed them at a high-throughput level. We dispense organoids with multidrop, and we can plate 40–50 384-well plates to perform drug screens in organoids. We’ve incorporated this in the use of liquid handlers and also imaging platforms.
Any high content screening microscope systems can also be used to image organoids and do image analysis. Cell viability is still the main readout for experiments in the oncology area, but we are looking for imaging readouts as we do with the swelling assay. The forskolin-induced swelling assay is also presented in the manuscript mentioned previously, is the readout we do for measuring CFTR restoration.
Where do you see the capabilities of your organoid models in 5 years’ time? Do you think they will become even more representative or do you see them being employed in new areas?
In 5 years, personalized medicine with organoid technology will be a reality. We will be able to assess what drug a cystic fibrosis patient should take using an organoid assay – our team is working on that now. Currently, we are already running organoid based clinical trials for cystic fibrosis patients. In parallel, we are working to move this application of organoids for personalized medicine and patient stratification for clinical trials in oncology for different types of tumors.
Can you tell me more about how you would employ organoids for cystic fibrosis patients?
We are now working on assay standardization and validation to get approval from the right entities (US FDA, EMA) to implement an HUB organoid-based test for a cystic fibrosis diagnostic, as we have already proven the predictive value of organoids in retrospective clinical trials. An organoid generated from a cystic fibrosis patient can predict if a patient will benefit from a treatment.
In individual cases in the Netherlands, for patients who’s lives are at risk and have no other alternative, their organoids have been generated, and if, in a FIS assay, the organoids have responded to a treatment, these patients have been given access to the tested treatment.
Is there anything else you’re working on at the moment that you’re particularly excited about?
We also have singular patient examples and corroboration from other research studies about organoid predictive value in oncology. For example, for colorectal cancer, organoids can predict patient response to treatment. We are validating this data by analyzing large numbers of patients and comparing patient and organoid data.
Apart of all the efforts for proving organoid clinical validation; the validation of the positive predictive value of organoids in oncology, we are also developing what we could call the 2.0 organoid model. We are combining organoids with other relevant cell types, such as immune cells or microbes to develop organoid models that better represent the pathophysiology of more complex diseases, such as inflammatory diseases, which will allow the testing of the efficiency of drugs in a patient-relevant model.
We are also hugely aware of advances in immune-oncology right now, and the relevance of combining tumor organoids with different cell types like T-cells or macrophages. So, we are developing more complex assays where we can assess immuno-oncology drugs in a relevant clinical model.