Visium Spatial Gene Expression technologies: an interview with the expert

 Spontaneous Russell (left) is a Staff Product Manager for Visium Spatial Gene Expression technologies from 10x Genomics (CA, USA). She has spent the last 20 years leading product development and product marketing efforts for life-science and diagnostic research tools. Most recently, she has been a core member of the product development team for the Visium CytAssist platform, designed for expanded sample access in spatial discovery with formalin-fixed, paraffin-embedded (FFPE) tissues.
Spontaneous Russell (left) is a Staff Product Manager for Visium Spatial Gene Expression technologies from 10x Genomics (CA, USA). She has spent the last 20 years leading product development and product marketing efforts for life-science and diagnostic research tools. Most recently, she has been a core member of the product development team for the Visium CytAssist platform, designed for expanded sample access in spatial discovery with formalin-fixed, paraffin-embedded (FFPE) tissues.
In this expert interview, she details how spatial technologies, specifically the Visium CytAssist platform, are revolutionizing translational research.
What difficulties are scientists facing in translational research at the moment and how can spatial technologies address these difficulties?
Tissue samples are a vast resource for translational research and valuable transcriptomic data lies within. Histology and immunofluorescence have delivered useful insights, but the gene expression profiles have remained difficult to fully understand within the tissue context. With spatial technologies, it is finally possible to resolve an incredible wealth of morphological and gene expression information that has been hidden inside of these tissues.
How does the Visium CytAssist platform work?
In the Visium CytAssist workflow (Fig.1), sectioning, deparaffinization, staining and imaging (H&E or IF) take place on a standard glass slide. After probe hybridization, two standard glass slides and a two–Capture Area Visium slide are placed in the CytAssist instrument so that the tissue sections on the standard slides can be aligned on top of the two Visium Capture Areas. Within the instrument, a brightfield image is captured to provide spatial orientation for data analysis, followed by hybridization of transcriptomic probes to the Visium slide. The remaining steps, starting with probe extension, follow the standard Visium for FFPE workflow outside of the instrument.
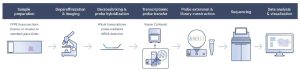
Figure 1. Visium CytAssist workflow from 10x Genomics.
How is this platform an improvement on previous spatial gene expression technologies?
The Visium CytAssist is a compact instrument designed to simplify the Visium workflow by facilitating the transfer of transcriptomic probes from standard glass slides to Visium slides, enabling spatial profiling insights to be gained from an expanded range of FFPE samples.
What challenges did you face when developing the Visium CytAssist platform?
When we set out to develop CytAssist, there was a huge range of features we considered incorporating. One of the biggest challenges for the team was striking a balance between the right kinds of features for the system. It was important for us to ensure that CytAssist would be accessible to a wide range of users, so keeping the system small and affordable while maximizing flexibility was a priority. Ultimately, we are very proud and excited about what we were able to deliver – a flexible, accessible, powerful platform for current and future Visium assays.
What are your top tips for using the Visium CytAssist platform?
Visium CytAssist is designed to bring the power of spatial transcriptomics to a broad range of samples through added flexibility in the processing workflow. Users of CytAssist can leverage pause points in the workflow to support more collaborative workflows that might involve sample transport and/or multiple handoffs of a sample along the journey from tissue block to spatially resolved gene expression data at the whole-transcriptome level.
How do you see this platform developing in the future and what do you imagine we can achieve with spatial technologies?
10x Genomics has established CytAssist as the platform for future spatial discovery applications based on Visium technology. We launched Visium CytAssist in 2022 with FFPE compatibility, and recently added support for fresh frozen samples. Our next key application is simultaneous gene and multiplexed protein detection, bringing spatial proteogenomics to the CytAssist platform.





