Sequencing reveals new cell types for the BRAIN initiative
One of the top priorities of the BRAIN initiative is identifying and providing experimental access to the different cell types in the brain, as well as determining their roles in health and disease. Different research labs are whole-heartedly embracing this challenge, with the help of innovative single-cell sequencing approaches.
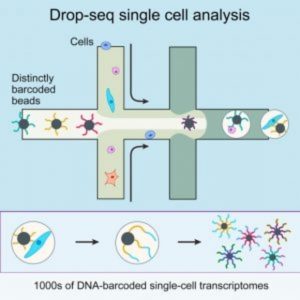
A diagram of Drop-Seq (2).
With nearly 100 billion neurons and 100 trillion connections, the human brain remains one of the greatest mysteries in science and one of the greatest challenges in medicine. Despite the many advances in neuroscience in recent years, the underlying causes of most neurological and psychiatric conditions remain largely unknown. To develop effective treatments, researchers need a more complete arsenal of tools and information for understanding how the brain functions in both healthy and disease states.
To address this challenge, former President Barack Obama launched the BRAIN (Brain Research through Advancing Innovative Neurotechnologies) Initiative in April 2013. This initiative scales from atoms to thoughts and behavior, linking what is known about single cells and subcellular activities to whole-brain function.
In response to this challenge, the National Institutes of Health (NIH) convened a working group to develop a rigorous plan for achieving this scientific vision. The BRAIN 2025 report released in June 2014 proposes how the initiative should develop over a 10-year period beginning in fiscal year 2016, with a primary focus on technology development in the first 5 years, shifting in the latter years to a focus on integrating technologies to make fundamental new discoveries about the brain.
Census of Cell Types
“One limitation is that we so far lack a comprehensive parts list of all the neurons that comprise the brain and a wiring diagram of how they connect to each other,” said John Ngai from the University of California, Berkeley. “How do these different cell types work together to serve the functioning of this exquisitely complex biological computer?”
With support from the BRAIN Initiative, Ngai and his team are working on classifying cortical neurons using single-cell transcriptomics. To understand what makes neurons distinct, they are exploring one major type of mouse brain cell, pinpointing the genes responsible for differentiation into cell subtypes, and testing whether each subtype has unique functions using a new technique that labels cells with tagged genes.
“We can use this information to identify novel cell types that previously eluded detection, simply because we did not have knowledge of markers that otherwise could be used to identify them,” Ngai said. “Ultimately, we can use single-cell transcriptomics as a foundation to acquire information about a cell’s location in the brain, morphology—including its connectivity with other neurons in the brain—and physiology, which together should provide all the information needed to identify, classify, and characterize every cell type in the mammalian brain.”
As reported in the June issue of Cell Stem Cell, Ngai and his team developed an integrated approach using in silico analysis of single-cell RNA sequencing data and in vivo lineage tracing—a technique for permanently labeling the descendants of a targeted cell. With this approach, the researchers illuminated the cell fate potentials of individual olfactory stem cells and the locations of branchpoints in the olfactory lineage trajectory at a level of detail not possible by either in vivo lineage tracing or single-cell RNA-sequencing alone [1].
“For the first time, we are now able to identify cell types and cell states within a heterogeneous population of cells without any a priori knowledge of what distinguishes one type or state from another,” Ngai said. “We literally caught cells in the act of choosing one cell fate over another and identified some of the genes driving such decisions.”
This combined analysis identified the trajectories that produce major cell types in the olfactory epithelium, and it serves as a model for elucidating complex lineage trajectories in other stem cell niches. According to the authors, detailed lineage trajectory maps are necessary for identifying the regulatory networks that govern the cell fate transitions underlying tissue maintenance and regeneration and are essential for designing strategies to manipulate cells for therapeutic applications.
Large-Scale Single-cell Analysis
On the opposite coast, at Harvard University, Joshua Sanes and his team are also using single-cell transcriptomics, focusing on new methods of genetic screening to comprehensively catalog and distinguish different kinds of cells across species and brain regions. “Our BRAIN grant was designed to compare various single-cell methods, using the retina as the main model, with the aim of then using the winner to generate a full cell atlas,” said Sanes.
In May 2015 in Cell, Sanes and his colleagues described Drop-seq, a strategy for quickly profiling thousands of individual cells by separating them into nanoliter-sized aqueous droplets, associating a different barcode with each cell’s RNAs, and sequencing them all together [2]. Drop-seq analyzes mRNA transcripts from thousands of individual cells simultaneously while remembering each transcript’s cell of origin. The researchers analyzed transcriptomes from 44,808 mouse retinal cells and identified 39 transcriptionally distinct cell populations, creating a molecular atlas of gene expression for known retinal cell classes and novel candidate cell subtypes.
“Drop-seq allowed transcriptomic analysis of tens of thousands of cells for the first time, by bringing the monetary and labor costs down by orders of magnitude,” Sanes said. “In fact, this paper may have reported as many single-cell transcriptomes as had been reported in all previous studies put together. The methodological innovation wasn’t just hardware though—improved bioinformatics methods were needed to process and comprehend the massive data sets.”
In another study, published in August 2016 in Cell, Sanes and his team developed and applied an integrated strategy for building a comprehensive validated atlas of cell types [3]. Using Drop-seq and optimized computational methods, the researchers studied a diverse class of neurons—mouse retinal bipolar cells. “Even though neuronal types in the retina have been studied for nearly 150 years, our work has identified multiple novel types,” said first author Karthik Shekhar, a postdoctoral associate at the Broad Institute of MIT and Harvard.
From a population of approximately 25,000 bipolar cells, the team derived a molecular classification system that identified 15 types, including all cell types observed previously plus 2 novel cell types. “We are close to completing the atlas of neuronal types for the retina, which will be the first of its kind in the mammalian brain,” said Shekhar.
According to the authors, this work provides a systematic methodology for achieving comprehensive molecular classification of neurons, identifies novel neuronal types, and uncovers transcriptional differences that distinguish cell types within a class. With the advance of multiplexing technologies and decreasing sequencing costs, future studies will be able to profile larger numbers of cells at greater depth.
“I believe that inability to comprehensively classify neurons is a major bottleneck to understanding neural function and dysfunction throughout the brain,” Sanes said. “I hope that success in the retina would allow us to optimize a pipeline that could then be applied to less tractable regions such as the cerebral cortex.”
According to Sanes, this effort is essential for finding out how circuits malfunction in brain diseases. “For example, some disorders may seem especially mysterious because they affect just a small number of the many types of neurons or glia in the brain, making it nearly impossible to track down the needle in the haystack,” he said. “As we classify neurons,and develop ways to mark and even purify them, we will be able to ask whether there are particularly vulnerable types.” In fact, their own work on the retina has already exposed some cell types that are more resilient to nerve injury than others.