A peek behind the paper: Pooled CRISPR screens with imaging on microraft arrays reveals stress granule-regulatory factors

Take a look behind the scenes of an article recently published in Nature Methods entitled ‘Pooled CRISPR screens with imaging on microraft arrays reveals stress granule-regulatory factors’, as we invite co-First Authors, Emily Wheeler and Anthony Vu, as well as Senior Author, Gene Yeo, to shed some light on their recent research.
Please can you give us a short summary of the method presented in your Nature Methods article, “Pooled CRISPR screens with imaging on microraft arrays reveals stress granule-regulatory factors”?
In this work, we developed an easy-to-use workflow that combines pooled CRISPR screens with high-content imaging and microraft-array technology to screen image-based phenotypes. We named the approach CRaft-ID (CRISPR-based microRaft, followed by gRNA identification). This method is inexpensive, employs standard laboratory equipment, and can be adapted to investigate a wide range of cellular and subcellular phenotypes.
As a proof-of-concept, CRaft-ID was utilized to screen a pool of cells infected with a lentiviral CRISPR library to identify genes that affect the abundance of stress granules. Stress granules are cytoplasmic RNA–protein aggregates that form under stress and are best analyzed using microscopy. This is a challenge for pooled CRISPR screens, which are limited to standard readouts that include survival or fluorescent markers for sorting. In order to perform a pooled CRISPR screen with an imaging readout, we customized the microraft array technology to easily culture thousands of distinct cell colonies in a shared media reservoir. We modified the microrafts by fixing a glass slide to the bottom of the array to enable, for the first time, high-resolution automated confocal microscopy across the entire array. To analyze the hundreds of thousands of microwell images, we developed image processing algorithms to identify colonies with our desired phenotype of reduced stress granule formation. Microrafts containing these colonies were then isolated, allowing us to identify and validate known and novel factors that affect the abundance of stress granules. This highlights the applicability of our approach to facilitate image-based pooled CRISPR screens.
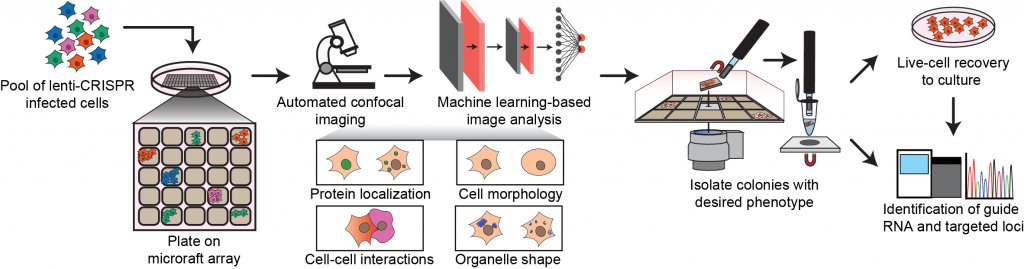
The CRaft-ID workflow.
What scientific problem inspired you to develop this new method?
Pooled CRISPR screens have transformed the life science community as an extremely powerful tool to identify genetic factors that are critical for a selected phenotype. Conveniently, this approach allows us to deplete thousands of genes simultaneously in a single sample, but until now we have been limited in microscopic visualization of that pool of cells. By modifying the microraft arrays with a glass back, we demonstrate a high-throughput method to screen a pool of genetically modified cells by an image-based phenotype, and importantly, provide a means to easily isolate colonies of interest for follow up studies.
Like this? Get in touch to publish your own Peek Behind the Paper now!
What impact do you hope it will have on laboratory researchers?
CRaft-ID will provide an inexpensive and accessible means for researchers to easily perform image-based pooled CRISPR screens in any standard laboratory setting. In contrast to current image-based screens that are performed in array format, our workflow does not require specialized robotic equipment for the manipulation of thousands of individual wells. The flexible platform is compatible with any CRISPR library, a variety of cell types, and countless cellular or subcellular phenotypes and therefore can be easily adapted by any researcher wanting to systematically annotate gene function with their phenotype of interest.
The technique appears relatively simple, with the software freely available online. Do you have any tips or tricks for others looking to utilize this method?
The microraft arrays behave like standard tissue culture plates, and therefore can accompany a wide range of cell types. However, nondividing cells are not suitable for direct plating onto the platform, where colonies are required for imaging and genomic DNA isolation. For such cases, we recommend users plate stem cells or intermediates with proliferating cells onto microraft and continue differentiation after colony formation.
Given that the penetrance of genome editing in cells transduced with lentiCRISPR guides can be highly variable, high coverage of a pooled CRISPR screen is recommended to increase the likelihood of capturing a knock-out for a selected phenotype. In our screen, we designed 10 unique sgRNAs targeting each gene and screened 10x as many cells as guides in the library, resulting in ~100 opportunities to phenotype each gene in the library. With this coverage, we are able to identify both novel and known genetic regulators of stress granule abundance. However, if higher coverage is desired, users should employ a smaller, focused library of sgRNAs or include more microraft arrays for imaging.
Before performing a screen with CRaft-ID, please read through this detailed step-by-step experimental protocol available online.
Critical steps in the protocol include:
- Centrifuging cells plated on microrafts in a swinging-bucket centrifuge to minimize the likelihood of cells adhering to microwell walls, resulting in out-of-focus colonies when imaging.
- Before plating onto microrafts: thoroughly dissociate trypsinized cells by repeated pipetting, strain the cell suspension, and plate at recommended cell density to reduce colony doublet formation.
- After isolating microwell with target cell colony, immediately proceed to DNA extraction with Quick Extract Buffer prior to storage at -20°C to increase the efficiency of genomic DNA extraction.
- Perform DNA extraction and first PCR reaction in pre-AMP PCR clean space to avoid library contamination.
What equipment would a researcher looking to utilize your method need to have to hand?
The CRaft-ID workflow was designed to be widely accessible by employing standard laboratory equipment, CRISPR libraries that do not require modifications, and commercially available materials. Microraft arrays can be purchased through Cell Microsystems (NC, USA) and will require the simple modification of securing a 1-mm-thick glass slide to the bottom of the array before imaging on any standard automated confocal microscope. Our CRaft-ID software used to process and analyze the acquired images is available in an open-access GitHub repository and can be executed on any conventional workstation. Finally, users will need a motorized microneedle device and a hand-manipulated magnetic wand to isolate the magnetized microrafts containing candidate cells for PCR-based sequencing. Both the microneedle device and magnetic wand are commercially available through Cell Microsystems.
What are you hoping to do next in this area?
After successfully identifying and validating novel and known regulators of stress granule formation, we will use CRaft-ID to discover genes critical for resolving stress granules after stress removal. Defects in stress granule assembly and clearance are firmly linked to neurodegenerative diseases and cancer, and therefore identification of genes essential to stress granule dynamics could reveal novel disease-relevant therapeutic targets. A unique feature of our image-based phenotypic screen is the ability to perform live-cell imaging of fluorescently labeled subcellular proteins followed by live-cell capture. By using an engineered cell line expressing a fluorescently labeled stress granule protein, we can now visualize real-time dynamics of stress granule behavior from our pool of lentiCRISPR-infected cells.
We also hope to expand the utility of CRaft-ID to other measurable subcellular phenotypes, including intracellular-RNA localization. Spatial and temporal regulation of RNA is critical for many cellular functions including cell motility, polarization and development, and therefore we aim to use our platform to determine which RNA-binding proteins are involved in the localization of mRNA transcripts critical for biological processes. Our approach is broadly applicable to many settings and we are excited to use it across new phenotypes that can be analyzed by advanced-microscopy technology and image-analysis algorithms.
PUBLISH YOUR OWN PEEK BEHIND THE PAPER NOW
 Gene Yeo is a Professor of Cellular and Molecular Medicine at the University of California San Diego (UCSD; CA, USA), a founding member of the Institute for Genomic Medicine and member of the UCSD Stem Cell Program and Moores Cancer Center. Yeo has a BSc in Chemical Engineering and a BA in Economics from the University of Illinois, Urbana-Champaign (USA), a PhD in Computational Neuroscience from Massachusetts Institute of Technology (USA) and an MBA from the UCSD Rady School of Management. He is a computational and experimental scientist who has contributed to RNA biology and therapeutics. His primary research interest is in understanding the importance of RNA processing and the roles that RNA binding proteins (RBPs) play in development and disease.
Gene Yeo is a Professor of Cellular and Molecular Medicine at the University of California San Diego (UCSD; CA, USA), a founding member of the Institute for Genomic Medicine and member of the UCSD Stem Cell Program and Moores Cancer Center. Yeo has a BSc in Chemical Engineering and a BA in Economics from the University of Illinois, Urbana-Champaign (USA), a PhD in Computational Neuroscience from Massachusetts Institute of Technology (USA) and an MBA from the UCSD Rady School of Management. He is a computational and experimental scientist who has contributed to RNA biology and therapeutics. His primary research interest is in understanding the importance of RNA processing and the roles that RNA binding proteins (RBPs) play in development and disease.
 Anthony Vu is a PhD student in UCSD’s Biomedical Sciences program. Prior to joining the program, Vu received a joint BS/MS degree in Biology from UCSD where he completed his master’s thesis work in Fred Gage’s lab at the Salk Institute (CA, USA) studying the molecular mechanisms by which critical neuron-specific gene repressor REST regulates neuronal development. In the Yeo lab, Vu is currently developing high-throughput genetic screens to study the mechanisms of protein aggregation underlying neurodegenerative disease to identify potential therapeutic targets. He is also interested in designing new tools to characterize the spatial and temporal regulation of RNA-protein complexes critical for neuronal function and health. Vu is a NSF GRFP fellow and an ARCS scholar since 2019.
Anthony Vu is a PhD student in UCSD’s Biomedical Sciences program. Prior to joining the program, Vu received a joint BS/MS degree in Biology from UCSD where he completed his master’s thesis work in Fred Gage’s lab at the Salk Institute (CA, USA) studying the molecular mechanisms by which critical neuron-specific gene repressor REST regulates neuronal development. In the Yeo lab, Vu is currently developing high-throughput genetic screens to study the mechanisms of protein aggregation underlying neurodegenerative disease to identify potential therapeutic targets. He is also interested in designing new tools to characterize the spatial and temporal regulation of RNA-protein complexes critical for neuronal function and health. Vu is a NSF GRFP fellow and an ARCS scholar since 2019.
 Emily Wheeler received her PhD in Biomedical Sciences from UCSD. Her thesis work in Yeo’s lab focused on using computational and molecular biology tools to uncover how protein—RNA interactions in cells play a role in cancer and neurodegeneration. Specifically, her work included uncovering novel RNA-binding proteins that are essential to form protein aggregates in motor neurons that cause cell death in amyotrophic lateral sclerosis. Additionally, she developed computational methods to determine how point mutations in splicing factors that occur in blood cancer alter their interaction with cellular RNA. She is a NSF GRFP fellow and an ARCS scholar since 2017. More recently, Wheeler joined Zuzana Tothova’s lab at the Harvard Medical School (MA, USA) as a postdoctoral fellow where she is studying the role of mutations in cohesion proteins to drive clonal hematopoiesis and blood cancer.
Emily Wheeler received her PhD in Biomedical Sciences from UCSD. Her thesis work in Yeo’s lab focused on using computational and molecular biology tools to uncover how protein—RNA interactions in cells play a role in cancer and neurodegeneration. Specifically, her work included uncovering novel RNA-binding proteins that are essential to form protein aggregates in motor neurons that cause cell death in amyotrophic lateral sclerosis. Additionally, she developed computational methods to determine how point mutations in splicing factors that occur in blood cancer alter their interaction with cellular RNA. She is a NSF GRFP fellow and an ARCS scholar since 2017. More recently, Wheeler joined Zuzana Tothova’s lab at the Harvard Medical School (MA, USA) as a postdoctoral fellow where she is studying the role of mutations in cohesion proteins to drive clonal hematopoiesis and blood cancer.