A prime time for genome editing
A novel genetic editing technique, prime editing, has been developed that improves on existing CRISPR—Cas9 techniques, producing higher precision and a wider selection of applications, potentially enabling researchers to correct 89% of established pathogenic genetic variants.
Recent research from the Broad Institute of MIT and Harvard (MA, USA) has dramatically advanced techniques in gene editing first established by the discovery of CRISPR, creating a new technique coined ‘prime editing’. The technique infers a higher level of precision to the gene-editing technique while potentially enabling researchers to rectify 89% of established pathogenic genetic variants.
Led by first author Andrew Anzalone and senior author David Liu, the team developed prime editing by modifying the existing CRISPR—Cas9 system to create a completely new way of editing DNA.
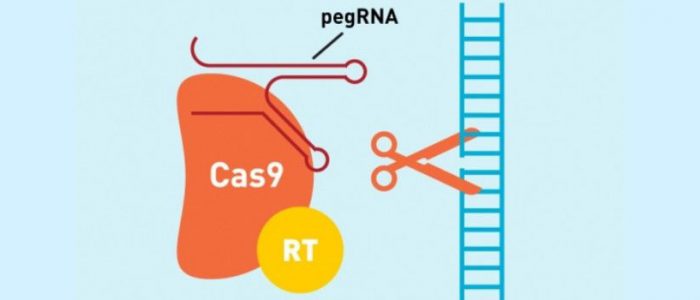
Prime editing is directed to the target site by a new kind of RNA, referred to as prime editing guide RNA (pegRNA). This pegRNA directs a catalytically impaired Cas9 protein, fused to reverse transcriptase to the target site, where the modified Cas9 cuts just one DNA strand, as opposed to the double-strand break induced in traditional CRISPR—Cas9 gene editing.
The newly developed pegRNA not only directs the Cas9—reverse transcriptase complex to the target site but also contains additional nucleotides that encode the desired gene edit. Once at the site the reverse transcriptase transcribes this additional RNA sequence into the target site.
The team has applied this technique to correct the gene variants that lead to Tay Sachs disease and sickle-cell anemia in human cells, processes that require the specific deletion of four bases and the alteration of a thymine to an adenine, respectively, exemplifying the precision of prime editing.
“The beauty of this system is that there are few restrictions on the edited sequence. Since the added nucleotides are specified by the pegRNA, they can be sequences that differ from the original strand by only one letter, that have additional or fewer letters, or that are various combinations of these changes,” marveled Liu.
- Expanding the reach of gene editing
- CRISPR—Cas9 induced off-target mutations are rare
- Talking Tech News | Theo Roth on CRISPR, CAR-T and gene editing ethics
The malleability of prime editing has been demonstrated in the paper recently published in Nature, which reports effective edits in mouse neurons and human cells. These edits include the 12 possible combinations of single base replacements, DNA sequence insertions up to 44 bases long, deletions up to 80 bases long and even alterations that include combinations of each of these edit types.
A further benefit of the prime editing technique is the reduced incidence of ‘off-target effects’ that occur with traditional CRISPR—Cas9. Prime editing can even make single-nucleotide edits in gene sequences that are inaccessible to base editors.
“A major aspiration in the molecular life sciences is the ability to precisely make any change to the genome in any location. We think prime editing brings us closer to that goal. We’re not aware of another editing technology in mammalian cells that offers this level of versatility and precision with so few by-products,” stated Liu, further admiring the capability of prime editing.
The Broad team will continue to develop and test prime editing while exploring delivery mechanisms for the technique in animals, with the ultimate goal of translating it into a viable gene therapy.
To perpetuate further the study of prime editing and to elicit additional research into its development, the Broad team will make the technology freely available to academic and non-profit organizations. This will include the distribution of vectors via the non-profit organization Addgene.
In a field that was already considered to be advancing at an exciting pace, prime editing – much like CRISPR did seven years ago – has jumped ahead of the curve and now looks set take the limelight in gene editing for the foreseeable future.