Expanding the genetic alphabet to create new life forms
After 15 years of hard work, synthetic biologists successfully engineered a healthy semisynthetic organism. This new life form can indefinitely store increased genetic information using an alphabet consisting of six letters that form three base pairs.
Cas9-based editing system (1).
The genetic alphabet, which is conserved throughout nature, consists of four basic building blocks of DNA: adenine (A), guanine (G), cytosine (C), and thymine (T), which form the A–T and G–C base pairs that make up the rungs of the DNA ladder. But for more than 15 years, Floyd Romesberg of the Scripps Research Institute has been trying to expand this alphabet to increase the information storage potential of DNA.
This effort has now paid off. As reported in Proceedings of the National Academy of Sciences, Romesberg’s team engineered a healthy semisynthetic organism that can indefinitely store increased genetic information using an alphabet consisting of six letters that form three base pairs (1). According to the authors, this organism constitutes a stable form of semisynthetic life, laying the foundation for efforts to impart life with new forms and functions not found in nature.
“Our approach for generating a semisynthetic organism is thus far the only method. No other lab has demonstrated the replication of an unnatural base pair inside of a living cell,” said Yorke Zhang, a graduate student in the Romesberg lab and first author of the study.
Optimizing Semisynthetic Life
In 2014, the Romesberg lab developed an unnatural “X–Y” base pair that formed between synthetic nucleotides (2). “We created it by first spending about 14 years developing the unnatural base pair in vitro so that it was stable when incorporated into DNA and so that polymerases could copy DNA that contains the X–Y unnatural base pair,” Romesberg said. “This basically involved synthesizing different variants of the X and Y and analyzing how they affect the copying of the DNA. Once we found a variant of X–Y that was well-copied, we learned how to get them into a cell and found that the cell could copy X–Y in their DNA.”
Specifically, the researchers grew Escherichia coli in the presence of unnatural nucleoside triphosphates, which are the building blocks required for copying the unnatural base pairs. They also enabled the bacteria to import them by providing a nucleoside triphosphate transporter, resulting in the replication of DNA containing the unnatural base pair. However, the organism grew poorly due to expression of the transporter, and it was unable to retain the unnatural base pair with extended growth even under optimal conditions.
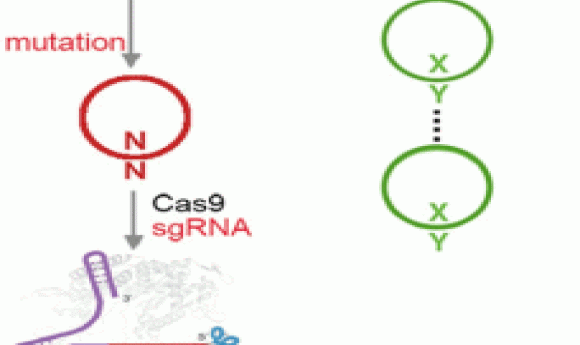
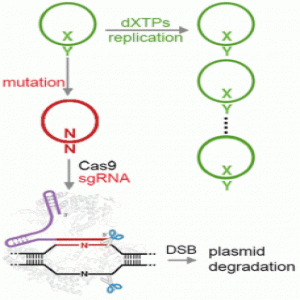
Addressing these shortcomings in the new study, the researchers engineered the transporter to be less toxic and chemically optimized the unnatural base pair, using a newer version of the base they denote as “Y” that is better than the previous version at being replicated and retained in the cells. They also harnessed the power of the bacterial immune response by using Cas9 to eliminate DNA that had lost the unnatural base pair.
“We employed the use of CRISPR/Cas9 as a mechanism to remove the natural sequences that arise from mutation of the unnatural base pair, showed that this ability was general across most sequence contexts, and ultimately demonstrated that CRISPR/Cas9 can enforce the retention of the unnatural base pair across many doublings,” Zhang explained. “This effectively immunizes the cell against loss of the unnatural base pair, demonstrating that CRISPR/Cas9 is an effective way to stabilize a semisynthetic organism.”
The result was a simplified and optimized semisynthetic organism that grows robustly and is capable of virtually unrestricted storage of increased information. According to Steven Benner, a scientist at the Foundation for Applied Molecular Evolution who was not involved in the study, this is an entirely different way to develop our understanding of life—synthesizing it from the bottom up.
“Here, the goal is to reproduce the properties that we value in life, but on different molecular platforms,” he said. “Floyd and his research group have grasped that grand challenge with both hands. This paper is an excellent example of what pursuits of that grand challenge can get by way of discovery.”
Making Better Drugs
According to Zhang, this approach has several advantages. “It doesn’t require drastic changes to the cell’s genome, requires only a few added components—the transporter, the CRISPR/Cas9 system, the unnatural base pair—to work, and is relatively inexpensive.”
The requirement for supplementation with unnatural triphosphates could also be considered a strength from a biocontainment standpoint. If the triphosphates are not provided, the cell is unable to replicate the unnatural base pair and will either die or mutate the unnatural base pair to a natural pair, preventing propagation of the synthetic component of the organism.
But the dependency on an external source of triphosphates places some constraints on the practical use of the semisynthetic organism. The triphosphates are not commercially available, so they must be made by custom synthesis, and they’re also degraded both externally in the media and within the cells, so the concentration of triphosphate available for replicating the unnatural base pair decreases as the cells grow if they are not supplemented with more. “The need for custom synthesis, and degradation constrains the ability to scale up growth of the semisynthetic organism, which is important if it’s ever going to be used to produce proteins containing unnatural amino acids,” said Zhang.
Moreover, the ability of the CRISPR/Cas9 system to eliminate natural mutations is very important for protein production. “For example, if and when the unnatural base pair is used to incorporate an unnatural amino acid into a therapeutic protein you are trying to manufacture, mutation of the unnatural base pair to a natural pair would result in fully natural proteins without the desired unnatural amino acid, and these would be considered contaminants in your production workflow,” Zhang said.
Although there are no practical applications of the semisynthetic organism at the moment, it could one day have important clinical implications. “We will have to demonstrate that the unnatural base pair can be transcribed and translated to incorporate unnatural amino acids into proteins for our approach to have utility, for example, incorporating unnatural amino acids into therapeutic proteins to make them better drugs,” Zhang said. “Since the stable storage of information, which this study demonstrates, is critical for faithful transcription and translation, this study provides a foundation for moving forward in that direction.”