Base editing for more efficient and representative organoids
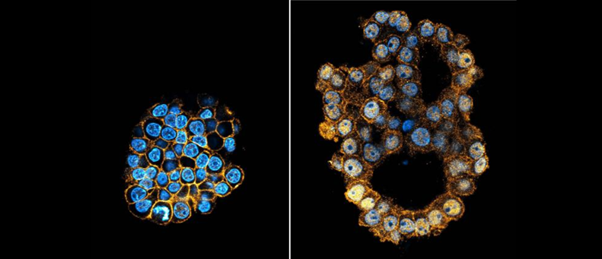
Using base editing, researchers have been able to establish tumor organoids with multiple mutations more efficiently.
A collaboration between researchers at the Organoid group at the Hubrecht Institute and the Princess Máxima Center (both Utrecht, The Netherlands), has successfully utilized base editors to integrate a suite of mutations into organoids derived from healthy tissue. Using this approach, the team has created more accurate models of liver, colorectal and endometrial tumors, producing new tools with which to establish novel therapeutics for cancer.
Tumor organoids can either be derived directly from patient tumor tissue or generated using cells from healthy tissue, which have been manipulated to include oncogenic mutations that are characteristic of specific cancers. However, many of the CRISPR/Cas9-mediated genome engineering methods used are limited to the knockout of genes and are less capable of inducing activating gene mutations. Further challenges arise with the off-target effects of these approaches and the laborious stepwise nature involved in establishing multiple mutations in the same cell means that there are also serious time constraints.
To address these issues, the researchers used a more recently developed technique: base editing. This approach couples a deaminase with the Cas9 protein, enabling it to induce a point mutation, without the deleterious double-strand break associated with other CRISPR-based approaches and with a higher degree of accuracy. To initially prove the function of this gene-editing system in organoids the team successfully introduced activating mutations into the CTNNB1 gene, which is often mutated in liver cancers, into healthy liver organoids using three separate base editors.
 New insights into HSV-1-induced encephalitis with cerebral organoids
New insights into HSV-1-induced encephalitis with cerebral organoids
A combination of anti-inflammatory and antiviral agents could improve treatment of encephalitis caused by HSV-1, according to new research utilizing cerebral organoids.
Commenting on the team’s observations of the cells after their mutation, first author Maarten Geurts stated that, “normally, this protein is located at the cell membrane, but in the organoids with mutations, we saw it in the nucleus and cytoplasm. This is because the protein is no longer broken down properly, so there is too much of it in the cells. What surprised us was that the exact effect was not the same for all CTNNB1 mutations. This gives us starting points to further investigate how exactly these mutations contribute to the development of liver cancer.”
To investigate the potential gain in efficiency that base editing could offer researchers, the team set out to induce several mutations into intestinal organoids. They successfully induced an activating mutation into PIK3CA, while simultaneously knocking out APC and TP53. With previous approaches, each additional mutation would require an extra 3 months of work. For a condition such as cancer, which typically involves multiple mutations, this development makes the generation of more accurate models with multiple mutations far more feasible.
With this newfound efficiency established, the team was then able to create a biobank of 128 intestinal organoid models containing different combinations of five gene mutations to mimic the various stages of colorectal cancer. Observing the differences between these organoids revealed a clear positive correlation between the number of mutations and the disruption to their morphology, which mimics the correlation observed in increasingly late-stage cancers to develop more chaotic shapes as they acquire more mutations.
Extolling the impact of this study and the utility of base editing, Geurts highlighted that the team had, “… made tumor organoids for three different organs in this study… We have thus shown that base editing makes it possible to make tumor models of different cancer types in a very efficient way. We have also shown that it is possible to incorporate up to five different mutations in one reaction. Since the efficiency of the reaction did not decrease, we may be able to increase this even further in the future. …. We are convinced that this provides us with a valuable tool for further research into the onset, development and treatment of various tumor types.”