New insights into the genetic basis of Parkinson’s

A new set of genes that contribute to Parkinson’s disease risk has been identified.
Why is it that some individuals with Parkinson’s disease (PD)-associated genetic variants develop the disease, while others do not? Are there other genetic factors at play? Now, researchers at Northwestern University (IL, USA) have identified a new set of genes implicated in developing PD, opening the door to new potential drug targets for treating this neurodegenerative disease.
Lysosomal dysfunction is a common feature of several neurodegenerative diseases, including PD. The lysosome is the recycling center of the cell, responsible for breaking down waste material, old cell components and other unwanted substances. Previous research has identified that mutations in the GBA1 gene result in decreased lysosomal glucocerebrosidase (GCase) activity – a key enzyme for lysosome function – and are linked to increased PD risk. However, incomplete penetrance of GBA1 variants suggests that other genes are also involved in PD manifestation. To investigate the other genes potentially associated with PD, the team conducted a pooled genome-wide CRISPR screen.
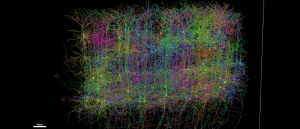
Largest brain wiring diagram generated from grain-sized sample
A global team has constructed the most detailed wiring diagram of a mammalian brain to date.
The team used a modern technology called CRISPR interference to examine the human genome. This gene-silencing technique works by blocking gene expression using an inactive form of the Cas9 protein, called dCas9, partnered with a guide RNA. The technique blocks gene expression without editing the DNA sequence; it can do this because the guide RNA can direct the dCas9 protein to bind – not cut – the promoter region of a target gene, which physically obstructs the RNA polymerase from transcribing the DNA.
“It also is possible to identify such genetic factors in susceptible individuals by studying tens of thousands of patients, which is challenging and costly,” explained corresponding author Dimitri Krainc. “Instead, we used a genome-wide CRISPR interference screen to silence each of the protein-coding human genes in cells and identified those important for PD pathogenesis.”
Examining genomes from the UK Biobank and AMP-PD, the team identified a group of 16 proteins – called Commander – that play a previously unrecognized role in delivering specific proteins to the lysosome for recycling. In people with PD, they observed loss-of-function variants in these Commander genes, demonstrating that a combination of genetic factors is implicated in PD manifestation and highlighting new potential drug targets.
“If Commander dysfunction is observed in these individuals, drugs that target Commander could hold broader therapeutic potential for treating disorders with lysosomal dysfunction,” Krainc concluded. “In this context, Commander-targeting drugs could also complement other PD treatments, such as therapies aiming to increase lysosomal GCase activity, as potential combinatorial therapy.”
Further research is needed to investigate the extent to which Commander proteins are involved in other lysosomal dysfunction-associated neurodegenerative diseases.