Accelerating antibody screening for viral threats
Antibody screening is an effective method for identifying disease. But can it keep up with viral epidemics?
Antibodies are key components of the human immune system. They are secreted by T cells and B cells to neutralise targeted pathogens. Historically, antibody screening has been utilized to uncover the antibodies and other immune cells found in a patient’s blood when infected with disease. This allows the appropriate therapy to be chosen for a patient.
They can also be utilized to discover new drugs. Antibodies make up a large amount of new drug discoveries because their inherent disease fighting ability makes them good candidates. Researchers from the University of Kansas (KS, USA) and the University of Texas (TX, USA) have pioneered a new method for rapidly screening a patient’s B cells. This has the potential to accelerate the discovery of new drugs for difficult-to-treat disease including viruses such as HIV, Ebola, Zika and Nipah.
VH:VL antibody screening ands rapid response
The new screen works by focusing on two specific regions of the antibody – the VH and the LH [1] — as author Brandon DeKosky explains:
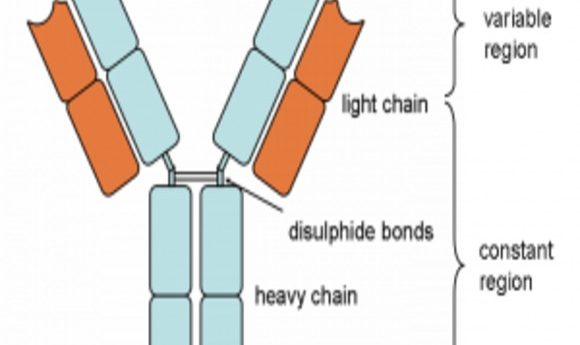
“Antibody molecules are encoded by B cells and are assembled from two different genes, called the heavy and light chains. The VH and VL portions – derived from the heavy and light chains, respectively – are the sections of an antibody gene that provide specific viral targeting. So, the VH and VL portions are the most important region to focus on for antibody screening and discovery.”
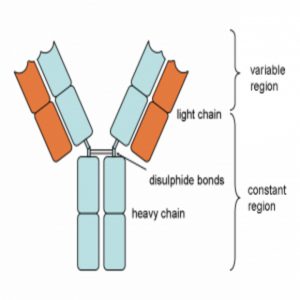
Diagram of an antibody showing the heavy and light chains from which the VH and VL portions are derived. Also shown are the variable and constant regions.
Previous attempts at rapid screening of antibodies have often failed due to the limitations of the technology. Previously, single-cell cloning had been used, which has a small sampling potential and high cost.
“Because antibodies are derived from two different genes – both of which are highly variable and contained within a single B cell – we need to perform single-cell manipulations en masse to recover the set of complete antibody genes. Traditional single-cell cloning is very expensive and time-consuming. The methods described here overcome those limitations and make it possible to screen millions of antibody-producing cells in a single experiment at an academic lab,” saidDeKosky.
However, these weren’t the only limitations.
“Non-natural gene pairing has been used in the past because it’s so much easier to do, but often the quality of antibodies discovered is not good enough for an effective drug, and the synthetic nature of those antibodies makes it difficult to understand the true human immune response. By maintaining native antibody gene pairings throughout our process, we identified antibodies as they occurred naturally. Along the way, we found extremely potent antibodies and learned more about the human immune response to vaccination and natural infection.”
The motivation for the study was the desire to understand human antibody response for vaccine and drug discovery. The group are hoping that their technique can be applied to those infected with HIV.
“Promising sources to discover new antibodies include donated blood samples from HIV patients with powerful immune responses against the virus, and also individuals who have received vaccines so that we can understand how those vaccines are working. When a potently neutralizing antibody is discovered, it can lead to new vaccine strategies and new therapeutic drug candidates.”
However, antibody screening has already been used for identification of viral immune response from HIV to pandemic viruses such as Zika and Ebola, and even a new virus that is currently terrorising India: the Nipah virus.
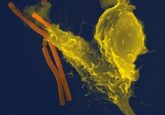
Scanning electron micrograph of HIV-1 budding (in green) from cultured lymphocyte. Multiple round bumps on cell surface represent sites of assembly and budding of virions. Source: CDC PHIL
HIV antibody screening
Traditionally, in order to carry out population screening for HIV, blood samples need to be taken from large amounts of people. An alternative to blood sampling is collecting oral fluid (OF). This can be advantageous because OF is easier to collect, does not require needles and is non-infectious. However a big disadvantage, that has held this method back, is the low concentration of antibodies in OF.
To effectively identify HIV infection through antibody screening using traditional methods, a high concentration of antibodies in the sample is necessary. However, a new assay developed by researchers at Stanford and Alameda County Public Health Laboratory (both CA, USA) has the potential to revolutionize antibody screening [2] as lead author Cheng-ting Tsai explains:
“There are a lot of populations you just can’t reach out to by blood tests. But if you were to do oral fluid, then all of sudden you open up a brand new population that was not otherwise accessible to you.”
The assay is based on antibody detection by agglutination-PCR (ADAP) technology. It does not look directly at the antibodies themselves due to their low concentration, but indirectly at the antibody’s effect. An antibody typically has two arms, when a pathogen such as HIV is detected the two arms bind to the virus as part of the immune response.
The researchers took a piece of DNA, split it in half and attached the DNA halves to bits of HIV. They then introduced the modified HIV to the OF sample. If the OF sample contained HIV antibodies then the two arms of the antibody molecule grab the modified HIV bringing the two halves of DNA together. The DNA molecule can then be easily sequenced.
The results of the study showed that HIV infection was easily detected in all 22 patients tested and Carolyn Baertozzi is hopeful that their method could supplant current antibody screening methods.
“Our hope is that we can get an earlier read than the present oral test because the sensitivity is better.”
Detection in pandemics
Zika
HIV has been at the forefront of virus research for the past 30 years. But in recent years new threats have arrived in the form of not well-understood pandemic viruses such as Ebola, Zika, and now Nipah. Speed is a key part of the response and scientists across the world have been working on these viruses; from identifying antibodies, to on-the-spot immunity tests, to indirect identification.
The Zika virus became a public health emergency when declared as such by WHO in 2016. Scientists quickly rushed to understand this disease and antibody screening formed a large part of their research.
A group of scientists from Humabs (Bellinzona, Switzerland) and Universities around the world set about identifying Zika antibodies from human B cells [3]. They took samples from four patients and were able to identify 100 Zika antibodies in 4 months. The study reported the first characterization of immune response to Zika infection and unearthed a potentially lethal interaction with another disease, dengue fever. Author Federica Sallusto explains the significance of the findings.
“In contrast to the broad cross-reactivity observed with antibodies, we found that the T cell response is to a large extent Zika-specific, a finding that may decrease the risk of Dengue infection in Zika-immunized individuals,”
This discovery accelerated new drug development and author Davide Corti, CSO and senior VP of Humabs, is excited by the prospects.
“I believe that our discoveries will also have a significant impact on the development of novel diagnostic to differentiate between a Zika or a Dengue infection”
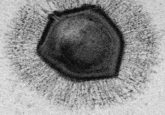
Ebola virus virion. Source: CDC PHIL
Ebola
The latest and most complicated Ebola outbreak began at a similar time to Zika. From 2014–2016 a lethal pandemic spread from West Africa to countries all over the world, bringing a huge amount of scientific focus onto the disease. In the following years, scientists have tried to develop diagnostic, management and surveillance tools for the disease.
Collaboration between scientists at Imperial College and UCL (both London, UK) has resulted in the development of a device that is able to diagnose immunity to Ebola by detecting IgG antibodies. It uses the latest technology for on-the-spot testing, working similarly to a pregnancy test and with the ability to be plugged into a smart phone for real-time results.
This could help identify patients in recovery and determining vaccination priorities for future outbreaks.
Nipah
Recent reported outbreaks of the Nipah virus in India have drawn attention to this newly emerging zoonosis. Its ability to infect both humans and animals with severe disease makes it a dangerous proposition for human health and agriculture. The disease is particularly known to infect pigs and has reservoirs in bats.
Research by scientists at Friedrich Loeffler Institut (Germany) has resulted in the development of an indirect ELISA for the detection of Nipah antibodies in pigs. The assay detects antibodies in blood samples from pigs and the group hopes the results may be used in agriculture for surveillance of infection for pigs and other livestock, helping to contain the risk of spread to human populations.