Accelerating biotherapeutic discovery with high throughput SPR
Antibodies represent the fastest growing class of drugs in the market today, with over 60 antibody drugs currently approved for use and more than 550 in clinical development1.
They are highly attractive as therapeutic agents as they have a naturally high affinity and specificity for their target antigen. These are desirable characteristics that enable an antibody to bind well to a single target, maximizing the desired biological effect, while minimizing off-target issues.
Antibodies can also be dosed at relatively low levels and administered less often (typically once a month) compared with small molecules, which is advantageous to the patient. In a recent development that demonstrates the power of antibody therapeutics in terms of patient convenience, TEVA Pharmaceuticals (Petah Tikva, Israel) has licensed AJOVY™ for migraine prevention, which only needs to be administered by injection once a quarter2.
These unique characteristics and therapeutic benefits represent lucrative opportunities for biopharmaceutical companies, however, the discovery and development process is currently labor-intensive and costly. Any method or technique that can reduce time and costs, while increasing the chance of finding and developing a successful therapeutic agent, is therefore much sought-after.
The Carterra® (UT, USA) LSA™ instrument performs high throughput surface plasmon resonance (SPR) for the characterization of antibodies. In the context of therapeutics, this technology has significantly impacted antibody screening, enabling higher-information-content assays to be conducted earlier in the research pipeline thereby streamlining lead selection.
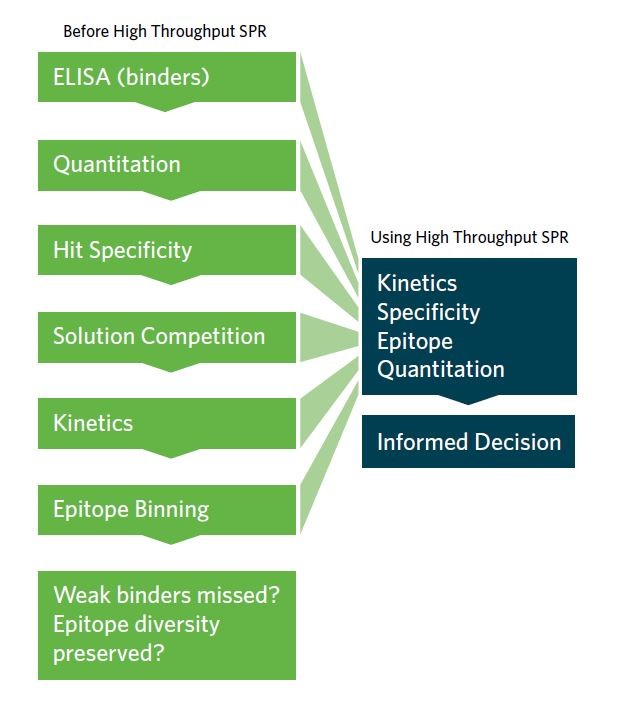
Figure 1. High throughput SPR revolutionizes antibody screening for drug discovery, allowing screening and characterization to be accomplished in the same step, at the very start of the discovery process.
Discovering a therapeutic antibody
The epitope specificity of an antibody is an innate property that cannot be designed rationally. Instead, an antibody with appropriate characteristics is uncovered through empirical investigation.
Since only the most promising leads are progressed into the clinic – typically a single antibody per program – it is essential that the right choice is made as early as possible. Spending time and resources chasing unproductive leads can result in a company losing its competitive edge in this fierce market.
Modern antibody generation technologies can produce prolific numbers of clones to feed discovery pipelines and expand the screening capacity. This increases the possibility of finding the next blockbuster drug that may be both transformative in treating a disease and differentiated enough to secure intellectual property, which can be considered the real estate of the drug industry.
Screening (or triaging) large libraries to identify antibodies with the most therapeutic potential involves detailed characterization of the molecular-level binding interactions between the target antigen and the antibody candidates. Ideally, this antibody screening should take place on the whole library at the earliest possibility to survey its functional repertoire. Assessing the epitope coverage of antibody libraries is also vital and, when combined with other data from orthogonal studies, can help to determine an antibody’s mechanism of action (MOA).
Antibody screening – then and now
Until recently, drug discovery scientists would commonly use high throughput but low-information-content assays (e.g. ELISA and FACS) as a preliminary screen to whittle down a large panel of antibodies to a manageable subset for low throughput but high-information-content SPR analysis. This approach is unsatisfactory because it underappreciates the full epitope and kinetic diversity of the original library, resulting in under-sampled pools and the possible selection of suboptimal clones that require extensive engineering. It is also likely to miss potentially high-value antibody drugs, or eliminate them from the sample pool prematurely.
High throughput SPR is now possible with the Carterra LSA, allowing screening and characterization to be accomplished in the same step. By expanding the capacity of SPR by an order of magnitude or more beyond that of other platforms (Fig. 2), Carterra’s LSA enables high-resolution and high-throughput interaction analyses to be performed on hundreds of antibodies in parallel. This allows SPR to shift upstream in the drug discovery process.

Figure 2. The Carterra LSA expands the capacity of SPR by an order of magnitude or more compared to other platforms. Kinetic data for 384 antigen:antibody interactions can be determined in parallel by the Carterra LSA, while other SPR systems can only produce kinetic data for eight (blue box) or 32 (green box) antigen:antibody binding interactions at a time.
The LSA also provides the advantage of minimal sample consumption and high assay detection sensitivity, making these assays amenable to early-stage research and discovery where large numbers of clones are available in low quantities.
High throughput SPR enables researchers to become better informed, earlier, because they can conduct detailed studies on the binding interactions between the target antigen and large numbers of antibodies.
Combining epitope binning and kinetic analyses
Knowing how an antibody binds its target influences its MOA, pharmacodynamics, and pharmacokinetics. SPR is the de facto biophysical method for generating binding kinetic and affinity data for antibody interactions but was traditionally low throughput and therefore used in secondary screening. The Carterra LSA is a disruptive platform because it enables SPR to be performed in expanded throughput, allowing it to fulfil a much-needed role upstream in preliminary screening, potentially obviating the need for ELISA.
Competitive binding assays on antibodies can be used to organize antibodies into epitope families or bins to help guide lead selection. The Carterra LSA transforms the binning paradigm making it possible to perform comprehensive epitope binning assays on large panels of antibodies quickly and with minimal sample consumption. For example, 384 antibodies can be epitope binned in a 348 x 384 assay format, generating data on more than 140,000 pairwise interactions in a single unattended run using less than 10 µg per antibody.
Binding parameters such as kinetics, affinity, and epitope specificity, provide researchers with a comprehensive information package that guides decisions on which antibodies to progress into further studies. Smart prioritization of resources is essential in drug discovery to quickly converge upon antibodies with the highest therapeutic value. With many companies often working on the same targets, there is a fierce race to the clinic and ultimately, to market approval.
Multi-parameter epitope binning and MOA
Elucidating the MOA of any novel drug, including therapeutic antibodies, is important for several reasons:
- Ability to make better-informed decisions that can impact both the safety and efficacy of a drug
- Help predicting a patient’s likeliness to respond to a particular treatment
- More-informed decisions on drug combinations to reduce the risk of resistance, likelihood of toxicity/side effects, and treatment failure
- Highlighting important molecular interactions and/or targets for further drug development
- Identifying novel MOAs that offer IP and commercial potential
Combining epitope binning data with orthogonal data in a process we refer to as multi-parameter epitope binning, can be used to construct a highly informative fingerprint for antibodies in a test panel and assess their therapeutic potential. The Carterra LSA platform delivers the throughput, data quality and multiplexing capabilities necessary to provide the comprehensive epitope binning studies for MOA fingerprints. Proprietary software enables the results from several orthogonal studies, such as neutralization, structural or mutagenesis data, to be merged with the epitope binning results, to visualize a detailed fingerprint (Fig. 3). This provides a functionally-relevant way of assessing the quality of an antibody panel to guide library-to-lead selection. Click here, to read a recent white paper from Carterra that goes into detail about the process of multi-parameter epitope binning for MOA determination.
18751
Figure 3. A network plot from a multi-parameter epitope binning experiment colored by orthogonal data. Neutralization of a Staphylococcus aureus virulence factor by the human antibody repertoire was investigated using high throughput SPR. Epitope binning was used to organize the antibodies into epitope clusters3. The researchers also performed cell-based functional studies to determine whether the antibodies blocked the target (IsdB) binding to its natural binding partner (hemoglobin) to neutralize S. aureus. Merging the neutralization and binning data revealed that hemoglobin blockade was restricted to two distinct bin clusters, namely bin C and bin P (red nodes), which suggested that two distinct MOA’s may be responsible for neutralization. This observation motivated further studies and was eventually confirmed by structural data.
Summary
Combining antibody screening and detailed characterization in the same step enables researchers to analyze libraries that are orders of magnitude larger than previously possible, obviating the need for preliminary screening by ELISA and facilitating the identification of near-optimal leads requiring minimal engineering. This expedites library-to-lead timelines and increases the chances that only clones with the highest potential for commercial and clinical success are progressed.
Drug discovery companies adopting this technology will benefit from faster and more highly informed lead selection, a reduced risk of failure, and the opportunity to expand and defend IP space – ultimately increasing the possibility of identifying the next Humira.
Carterra® is a new and innovative provider of technologies designed to accelerate and improve the discovery of novel therapeutic candidates. Carterra’s high throughput LSA™ instrument for monoclonal antibody (mAb) characterization delivers up to 100 times the throughput of existing platforms in 10% of the time while using only 1% of the sample. LSA combines patented microfluidics technology with real-time array Surface Plasmon Resonance (SPR) and industry leading data analysis and visualization software to revolutionize mAb screening. The LSA provides customers with mAb characterization throughput and functionality that matches the output from state-of-the-art antibody expression platforms. This enables all antibodies to be rapidly and comprehensively screened early in the discovery process so that unique epitopes and potential novel therapeutic candidates can be identified, while expanding and enhancing IP coverage.