Coming of phage: treating antibiotic-resistant infections with bacteriophages

© University of Leicester
Martha Clokie (left) is a Professor of microbiology at the University of Leicester (UK), where she runs the first Centre for Phage Research in the UK. Her research is focused on different aspects of bacteriophages – natural viruses that target specific bacterial groups – and how they can be harnessed to treat antibiotic-resistant infections.
In this interview from ELRIG Research & Innovation (20–21 March 2024; Manchester, UK), Digital Editor Beatrice Bowlby spoke to Clokie about phages’ diverse structure and function, the roles that both natural and engineered phages could play in overcoming the growing issue of antibiotic-resistant bacterial infections and what happens if bacteria become phage-resistant.
How do bacteria become antibiotic resistant and why has it become such a big problem?
Many people think resistance has come about solely because we use too many antibiotics. However, it actually goes back a long, long way. Bacteria live in very complex environments with many other species, making these spaces very competitive. The bacteria therefore send out compounds to kill the other bacteria, which is where most antibiotics we use are derived from. Because bacteria have been around for 3.9 billion years, they’ve developed defense strategies against these compounds. For example, if a compound blocks a certain pathway, the bacteria have another pathway, so they don’t need the original pathway or they modify the target site. Often, bacteria develop resistance via efflux pumps that allow them to pump the compound out straight away, or they can deactivate the compound as it comes in. They have many different strategies to become resistant, and for example, bacteria such as Pseudomonas – a highly problematic bacteria causing all sorts of infections in the lungs – can utilize all of these strategies, making it very difficult to treat.
Of course, it is true to say that the more we use antibiotics, the more we drive this resistance. Additionally, bacteria are good at exchanging information with each other that promotes resistance; the resistance can be encoded in plasmids for example, and those plasmids can go from one species to another and spread the resistance message.
Why are bacteriophages potentially good vehicles for treating antibiotic-resistant infections?
If you look at that sort of environment that contains the bacteria fighting each other, the other dominant player in terms of abundance is the bacteriophages, which infect and kill bacteria, profoundly shaping bacterial communities. They haven’t been investigated in great depth as antimicrobials because they were seen in the past as being too complicated. With an antibiotic, we could just utilize the one compound that one type of bacteria was putting out to kill another; we could purify that to make a nice, clean product that we could test to develop standard treatments and dosage. Additionally, antibiotics cast a wide net, perhaps killing many species and generally every type of variant within that species.
Bacteriophages are much more complicated. First, we must find the exact receptors on the bacteria that allow bacteriophage infection, and these are very specific. Some phages are so specific that they won’t even kill all the Staphylococcus, for example, they’ll kill only the ones with a particular protein sticking out with a particular modification on the side. This specificity makes it much more complicated to know which phages to use. You need better diagnostics to make sure that the correct phage is used for certain bacteria. However, specificity is good because if you have a gut infection and it’s caused by an E. coli, we can give you some phages that remove the E. coli and leave the rest of the microbiome intact. It’s this complexity that we now need in terms of treatment. The techniques we have available to us characterize phages well, which means we can identify and construct their composition, and ultimately engineer them.
This complexity and specificity certainly contributed to why phage technology never really took off in the Western world. However, phages give us a whole new way of killing bacteria. Phages that kill each bacterial species can be vastly different from each other; there is so much new biology to unravel, study and exploit, making phages vital for the next phase of infection treatment. In terms of understanding how to kill bacteria, it totally makes sense to study their smallest, most abundant and most effective enemies – bacteriophages – to learn how they do it.
What techniques do you use in your phage development research?
When I started in this field around 20 years ago, I gathered all the techniques I could because I was aware it had been such an unpopular area of science for so many years that all the people who developed them were retiring and leaving the field. With colleagues in Canada and Belgium, I edited four different volumes of protocols (see below) over a few years to try and compile all the techniques. We use a wide range of techniques because we’re trying to cover a lot of ground: from well-established ecological techniques – for finding the phages – to sampling and bringing them from the wild into a laboratory setting. Once in the lab, we purify the phages by identifying and growing them on specific bacterial hosts.
Currently, when producing phages, we use a range of specific bacterial hosts – the bacteria that we wish to identify phages for with the view to kill them in clinical/veterinary/agricultural settings. We grow the bacterial hosts, introduce phages, allow them to multiply and then separate them from their now lysed bacterial cells. One of the problems with this preparation is that it’s hard to collect those phages as they’re amongst bits of broken bacteria, which can be difficult to separate and can cause harm when introduced into animals. Now, people are developing new and better ways to purify phages so this challenge can be overcome. Another way to cultivate phages, called cell-free phage production, is being developed. This is based on the question: can we throw all the elements into a soup and create phages from that? This is just one of the exciting areas for the future of phage technology.
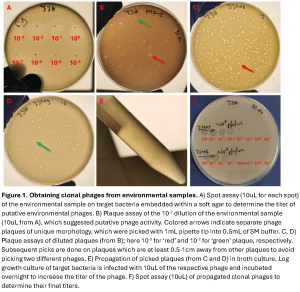
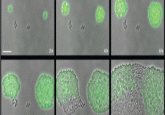
In terms of our most common flow of work, when trying to find phages, we must venture into the environments where those phages will likely be present and then grow them on a liquid exponential growth culture of cells. Often these can be environments such as sewage works, animal feces, rivers, soils or mud – anywhere where a diversity of relevant bacteria can be found in abundance. The next question to investigate is, can these now amplified phages kill the bacteria of interest? For this, we use a spot assay, which is when you allow the phage sample to interact with the bacteria and monitor the presence of lysis zones – these indicate the presence of a phage. Next, we scrape zones of lysis and put that scraping into another round of assaying, producing many different zones (see Figure 1, provided by Martha Clokie). We repeat this step multiple times because initially there could be more than one phage in the plaque; therefore, this acts as a purification step.
After this purification, we sequence the phage’s genome. It used to be that we would do pulse field gel electrophoresis, which reveals the size of the genome. But these days, the cost of sequencing has gone down so much that we use either Illumina-based or nanopore sequencing. At this point, we also want to establish the phages’ host range, so we do host range analysis by seeing if the phage can infect a representative diversity of bacterial cells. The reason why we sequence the phage genome is to make sure that they aren’t carrying any toxins or anything that might make the infection worse and to understand and classify the virus better. One problem we have had is that phage genomes are so diverse that they encode a huge number of ‘hypothetical genes’ – genes for which there is no known function. Therefore, we’ve been using AlphaFold to inform on the structure of proteins encoded by these hypothetical genes.
We also use transmission electron microscopy and sometimes, to understand our phages better, we examine phage transcriptomes. Sequencing the transcriptomes shows us how the phage overtakes the transcription and translation machinery of the bacteria. We also often carry out proteomic studies to identify the structural components of phages.
Another useful technique is epifluorescence, which we use to count the number of phage particles in samples. This is particularly useful if we are going to sequence viral metagenomes as it gives us confidence that we have suitably high numbers of viral particles present. For example, if we want to investigate the prevalence of phage in chronic obstructive pulmonary disease – a complex chronic disease that’s associated with flare ups in particular bacterial populations during exacerbation – we take a sputum sample from a patient, break down the mucus, stain the phage particles and count them using epifluorescence before sequencing that viral fraction.
Can you tell me about the phage research you’ve done specifically with Clostridium difficile?
I should have known when I started working on this organism – and I think the clue was in the name – how difficult it would be to work with. C. difficile is an obligate anaerobe in the gut, which many people carry. However, it becomes a problem when one goes to hospital and is given a broad-spectrum antibiotic, which decimates most of the gut community and allows C. difficile to run rampant. When this happens, C. difficile secretes toxins that destroy the tight junctions of the gut lining, which is why these individuals get diarrhea. It’s a disease that has persisted, remaining the biggest cause of infectious diarrhea in hospitals. Our goal was to find a lasting treatment, and since very few studies had explored phages for C. difficile before, I and a newly established team started to look for phages that infected and killed this type of bacteria. Eventually, after three years of searching, we found some phages.
We worked extensively to identify standard lytic phages that kill C. difficile, but this proved very difficult. I first tried sampling from patients because the dogma is (as I said above!) that where you find lots of bacteria, you’ll find the phages. However, we didn’t find any phages in these samples. Then, we moved to looking at samples from babies because it’s thought that they don’t have the receptors for toxins, and therefore, they have higher carriage rates of Clostridia than adults. We were able to isolate one effective phage, called R9, from my 9-month-old baby’s stool sample. However, our best phages actually came from marine estuaries. This is because if you dig deep enough, you hit anoxic mud, which is a natural environment for Clostridia species, and therefore their phages flourish and infect the bacteria.
From this work, I learned that it’s best to retrieve phages from healthy organisms, as it’s in these organisms that you find successful phages. This is the difference between phages and antimicrobials or drugs; phages are a part of the bacterial landscape. This means that we’re not introducing something totally different by using phages as treatments; our bodies are used to them. In this work, we brought C. difficile phages into cultivation, and this has facilitated our understanding of their potential treatment abilities. We have had a long-standing program since then of characterizing these C. difficile phages, working out which ones most effectively kill clinically relevant strains of this bacterium. We showed efficacy in different settings aimed at replicating how the bacteria naturally infect – in environments like artificial guts, insect infection models and a well-established hamster model. We still have ongoing research in this area.
Can bacteria become phage resistant and, if so, how will this affect the way that phage therapies are implemented and adapted?
When I first started working on phages, people used to say there were three ways that phage resistance evolved. One is that bacteria can change the surface of their cells to stop the phages from getting in. The second way is that they can alter their methylation; they can, for example, methylate their own DNA, which means that when the unmethylated phage DNA comes in, they can identify and chop up the phage DNA. The third way is called ‘abortive infection’ whereby the phage enters, and the bacterium commits suicide via a signaling cascade as an altruistic act to save the rest of the cells within that population. However, now I believe there are around 126 different phage defense systems that have been identified but not all of them characterized. Additionally, not all of them are present in every species.
Characterizing these defenses will be critical to deploying phages to treat disease, so a great deal of work is going into this area. There are multiple ways to approach the characterization of bacterial defense systems against phages. In Georgia, researchers co-evolve phage-resistant bacteria with phages, which works because phages naturally find a way around bacterial defense systems. By exposing phages to the bacterial pathways that they will be exposed to later on, they can evolve the necessary anti-defense strategies.
Another strategy for overcoming these bacterial defense mechanisms is one that encourages phage therapy and antibiotics to work in tandem. Researchers at Yale University (CT, USA) – Ben Chan and Paul Turner – found phages that bind to the efflux pumps of bacteria. This is clever because the only way those bacteria can become resistant against the phages is to mutate their efflux pumps, which means that they are then re-sensitized to the antibiotics. This strategy has been successfully implemented in a fairly large patient cohort, demonstrating that understanding resistance is key to developing phage products.
In terms of our next steps to implement phages as therapeutics, I think we have a real opportunity with phages to solve antimicrobial resistance. They have evolved multiple new ways to kill bacterial pathogens. We should understand their biology to develop them sensibly and to avoid all the mistakes that we made with antibiotics; we can approach phage therapy in an informed and regulated way.
I think we will have to adopt a new mindset – antibiotics were once seen as an essential way of removing all bacteria, including those that made us sick. This had a devastating impact on our microbiomes, and the overuse of antibiotics drove bacteria to become resistant, creating the landscape we have today where doctors are seeing increasing numbers of bacteria they can’t treat. Phages, in contrast, can be seen as the selective removers of bacterial species to improve one’s health. They are part of a natural process and will leave our beneficial microbiomes in place. Our ability to readily access sequence and other ‘omics data can help us in this quest.
The opinions expressed in this interview are those of the interviewee and do not necessarily reflect the views of BioTechniques or Taylor & Francis Group.