Expansion revealing microscopy exposes unseen cellular nanostructures

A technique called expansion revealing microscopy separates molecules in cells before fluorescence labeling, uncovering unseen nanostructures for the first time.
A research group at MIT (MA, USA) has developed a technique to separate molecules in a cell or tissue sample before fluorescently labeling them, enabling cellular structures to be visualized at a resolution of 20 nanometers.
The capabilities of conventional fluorescence imaging are often limited as antibodies need to be able to access the molecule of interest to bind a fluorescent label. As living cells are densely packed together some molecules are inaccessible to antibodies, which means they cannot be labeled or visualized.
To overcome this, the research group developed expansion revealing microscopy, which improves upon a previous technique they published in 2015 called expansion microscopy. In the earlier technique, an enzyme is used to expand samples; however, the enzyme cleaves proteins meaning that fluorescent labeling has to take place before expansion.
Now, the researchers have found a way of expanding samples that leaves the proteins intact so fluorescent labeling can be done after sample expansion. The new and improved method uses heat, instead of enzymes, to soften the tissue and results in a 20-fold expansion. Separating molecules in samples means antibodies can easily access and label proteins.
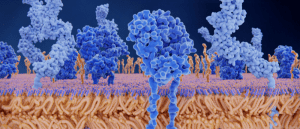 Preparing the troops: cryo-EM image provides insight into T-cell activation
Preparing the troops: cryo-EM image provides insight into T-cell activation
Researchers use cryo-EM to study how signaling events are activated following a T-cell receptor–antigen interaction.
By ‘de-crowding’ samples in this way, there is the potential for new cellular nanostructures to be uncovered, as demonstrated by the researchers who identified several cellular structures in synapses.
“This technology can be used to answer a lot of biological questions about dysfunction in synaptic proteins, which are involved in neurodegenerative diseases,” said Jinyoung Kang, one of the co-lead authors. “Until now there has been no tool to visualize synapses very well.” The researchers were able to label and identify seven synaptic proteins and visualized ‘nanocolumns’ in detail, which are made of calcium channels aligned with other synaptic proteins.
In addition to synapses, the researchers used the technique to image beta amyloid in brain tissue from mice, a peptide that forms plaques in Alzheimer’s patients. They found that the plaques form periodic nanoclusters, something which has never been seen before. They also observed that the clusters contain potassium channels and form helical structures along axons. “In this paper, we don’t speculate as to what that biology might mean, but we show that it exists,” said Margaret Schroeder, one of the co-authors of the paper, adding that this is just an example of new patterns that can be identified using this technique.
The researchers are now collaborating with other research groups to study different cellular structures including protein aggregates linked to Parkinson’s. They are also modifying the technique to be able to visualize 20 proteins at the same time and adapting it so it can be applied to human tissue samples.
Boyden concluded: “Time and again, you see things that are truly shocking. It shows us how much we are missing with classical unexpanded staining.”