Real-time microscopic video
Recent advances in microscopy methods have enabled visualization of “known” molecular processes in action.
In 1881, a time when anthrax killed millions of animals each year, Louis Pasteur hypothesized that an attenuated strain of anthrax bacteria could immunize cattle against the disease. In a dramatic public demonstration in Pouilly-le-Fort, France, Pasteur inoculated a group of farm animals with a low virulence anthrax bacteria strain, then two weeks later, injected them with highly virulent anthrax bacteria. He also injected a control group of non-vaccinated animals with the virulent strain. Hundreds of spectators, including scientists, politicians, veterinarians, and the general public gathered three days after the injections to witness his shocking findings: animals in the control group died, while the vaccinated animals remained healthy.
As Pasteur described in his report, “A large number of them—if not all—had been incredulous at the results of our [previous] treatments. Today, these individuals are the most enthusiastic disciples of our new doctrines ” (1).
The fact that Pasteur had to prove his hypothesis before witnesses to win over their confidence is not entirely surprising. Humans tend to believe what they see, so even scientists need innovative ways to convince the world and each other of their logical inferences.
Seeing is Believing
It is not surprising that microscopy, the most literal form of visual demonstration in science, has evolved remarkably over the years, branching into several application-based sub-types. For instance, development of fluorescence microscopy enabled researchers to visualize biomolecules tagged with fluorescent dyes. The compatibility of this method with living cells transformed it into a common lab-tool for visualizing cellular compartments and proteins within them. Next, researchers focused on improving the specificity of the fluorescent probes targeting the biomolecules of interest to ensure reliable data devoid of off-target signals.
According to Stefan Sarafianos from Emory University, in situ hybridization immunofluorescence, has improved tremendously over the years. Dramatic improvements in commercially available labeled probes have greatly enhanced the signal to noise ratio, enabling researchers to work on questions that seemed impossible only a few years ago. “It is a fantastic tool to communicate ideas that can be readily digested by people who are not necessarily experts,” said Sarafianos.
In their recent work, Sarafianos and his team visualized reverse transcription kinetics during HIV replication for the first time using an immunofluorescence based approach: multiplex immunofluorescent cell-based detection of DNA, RNA, and Protein (MICDDRP). Using this novel method, the team observed interactions of fluorescently labeled nucleic acids and proteins simultaneously, and discovered that reverse transcription begins in cytoplasm during early stages of infection (2).
AFM: Fingers Sense What the Eye Cannot See
Although recent superresolution microscopy methods have pushed the diffraction limits of resolution for fluorescence microscopy, it remains an indirect observation method relying on fluorescent probes. Thus, for direct visualization of molecular protein structures, researchers turned to non-optical microscopy alternatives.
One such technique, atomic force microscopy (AFM), consists of a sharp needle at the end of a flexible lever for scanning the sample surface. A laser beam is projected on the lever head, so sample roughness resulting in bending of the flexible lever deflects the laser beam, yielding a topographic map of the sample with <1 nm lateral resolution.
“AFM pictures molecules like one reads a Braille script. It is like a nano-finger that can push and pull molecules, manipulate them, measure elasticity of soft and harder materials,” explained Frederic Eghiaian from JPK Instruments, a leading AFM manufacturer. The sharper the needle apex, the better the lateral resolution, and that faster it can scan, yielding improved temporal resolution. Yet, researchers struggled for almost a decade to develop high-speed AFM (HS-AFM) to scan biomolecules in action without damaging them.
Finally, in 2010, Toshio Ando from Kanazawa University, Japan, successfully captured a video of a myosin motor walking on an actin filament using an HS-AFM that scanned 100X faster than usual AFM, as reported in their study published in Nature (3). “The group of Toshio Ando basically found all solutions which made such AFM experiments possible: they worked on all aspects, from the protocol of scanning, detection and signal treatment to the AFM probe itself and even the sample holder,” said Eghiaian.
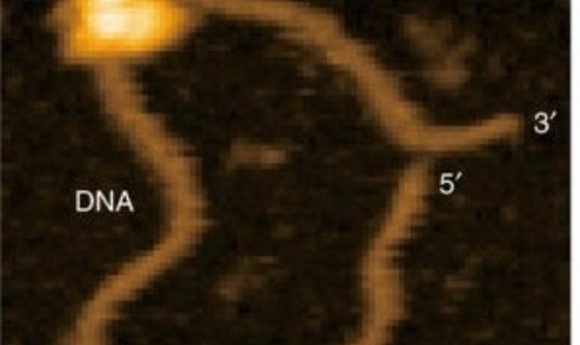
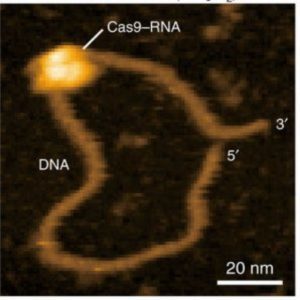
Recently, Ando and colleagues applied their HS-AFM technology to visualize the mechanism of the popular CRISPR/Cas9 machinery in real time (4). The CRISPR/Cas9 system consists of a guide RNA that directs the nuclease, Cas9, to specific DNA sequences, which the nuclease then cleaves. The video by Ando’s team clearly shows the Cas9 nuclease cleaving the DNA, providing the first visual confirmation of this mechanism.
Although the findings confirm a previously suggested hypothesis, the importance of these data cannot be underestimated, according to Eghiaian. “Did we really know the CRISPR mechanism?” he questioned, arguing that inferences based on structural and optical microscopy data, although solid, were still just the best available hypothesis. Ando’s team revealed minor details in the cleavage mechanism, such as fluctuation of a Cas9 domain between two conformations before adopting its catalytically active form.
“Such an observation is not only novel, but it is also different from what could be concluded from optical microscopy observations,” Eghiaian explained. “Seeing CRISPR at work should feel the same as learning about round earth at school, and then going to high altitude to witness the curvature of earth and realizing that earth really is round, and then one can get more precise mapping of earth, prediction of meteorological events, and so much more” he added.
Cryo EM: Powering the ‘Resolution Revolution’
AFM is excellent for imaging and analyzing physical properties of cells and single molecules, but it provides no information regarding their molecular composition. For many decades, structural biologists used X-ray crystallography to determine atomic and molecular structures of proteins. However, the sample preparation requires crstallization of proteins, and is thus unsuitable for several protein types that are difficult to crystallize. The introduction of cryo EM, which involves freezing biomolecules and imaging them using a non-damaging beam of electrons, provided a potential alternative.
In the beginning, however, cryo EM suffered from low resolution due to inefficient detectors that yielded blurry images due to movement of proteins in solution. Some researchers dismissed the method, terming it “blob-ology,” while others persevered. According to Edward Morris from the Institute of Cancer Research, London, the most significant breakthrough in cryo EM in the past few years has been improvement in the detectors. Researchers developed high speed cameras capable of capturing hundreds of images of the protein, which could be “un-blurred” using sophisticated new generation software to render 3D models of proteins at atomic resolution.
“This has been the biggest factor in making it possible to use the single particle cryo EM technique to get to atomic resolution and to see the detailed mechanism of protein complexes,” said Morris.
Recently, Morris and his team used cryo EM to image RNA polymerase III to investigate the transcription mechanism in detail. Their results showed for the first time how components of the RNA polymerase complex interact with one another (5). “We are not recapitulating something that was already known, there actually are novel observations for the most part,” Morris emphasized.
As RNA polymerase III, which transcribes non-coding RNA, is often disregulated in cancer cells, the teams hopes that the information from their study could help develop targeted cancer drugs in the future.
The Future is Interdisciplinary
The different microscopy methods are no doubt aiding tremendous scientific progress individually, but researchers believe that convergence of these methods will make them even more powerful.
Combining AFM with super-resolution microscopy techniques could help locate zones of interest for nanoscopic studies, according to Eghiaian. For instance, a researcher could spot his biological target using high-resolution fluorescence, and image or manipulate it in in vitro, ex vivo, or in vivo experiments using AFM, he explained.
On similar lines, Sarafianos suggested combining immunofluorescence with a powerful technique like cryo EM to pick the right cellular compartment using fluorescence, and then using cryo EM to zoom in on the specific part of certain cell at a precise location.
“Imagine you have a high-end telescope and you use it to go through the whole sky to find Jupiter. Well, you first need to know where Jupiter is to use your telescope,” Sarafianos explained.
In future endeavors, researchers hope to continue unraveling mechanisms of biological processes using novel state-of-the-art microscopy methods at the intersection of physics, chemistry, and biology. “Seeing is not just believing; it often yields more information than blind, indirect observation,” noted Eghiaian.