Stronger together: collective resistance to antibiotics
Refer a colleague
Bacteria susceptible to antibiotics get by with a little help from their resistant friends.
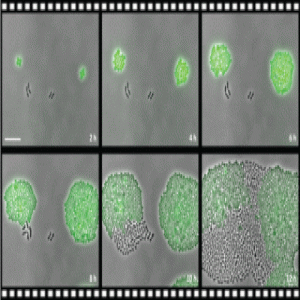
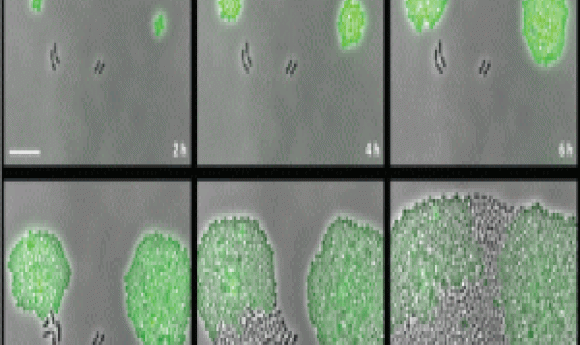
Susceptible Pneumococcus take advantage of the resistant strains around them to survive and proliferate when facing antibiotic wipe-out (1).
It is not over-dramatic to state that global antibiotic resistance is a health crisis, with the World Health Organisation declaring that it “poses a fundamental threat to human health, development, and security.” From the reporting of antibiotic misuse in animal feed to characterizing the mutations that allow resistance to develop, it regularly features in health and science news headlines. Now, new research published in PLoS One reports a novel non-inherited mechanism by which bacteria susceptible to antibiotics act as freeloaders—passively utilizing the resistance of surrounding strains to survive and proliferate.
“This work highlights why it’s so important to look at entire microbial contexts during infectious diseases, with multiple strains of bacteria generating a community of resistance, creating another potential complicating factor in antibiotic therapy,” said Robin Sorg from the University of Groningen in The Netherlands, first author of the study.
Sorg’s team focused on the opportunistic bacteria Streptococcus pneumoniae (Pneumococcus) and the antibiotic chloramphenicol, which is prescribed widely in low-income countries due to its broad spectrum and availability. The resistant strain of Pneumococcus they chose expressed chloramphenicol acetyltransferase (CAT), which provides resistance by preventing the drug from binding to bacterial ribosomes.
Using real-time single-cell analysis and luminescence, Sorg’s team confirmed that susceptible Pneumococcus bacteria grew in media supplemented with chloramphenicol when CAT-expressing strains were present. “We created a time lapse videousing GFP that nicely summarized this phenomenon of collective resistance,” stated Sorg. The CAT-expressing and susceptible bacteria, labelled in green and black respectively, survived the antibiotic medium, with the susceptible cells even eventually outgrowing their resistant counterparts.
Next, the researchers used ecological modelling to test the scope for co-existence between the CAT strain and susceptible Pneumococcus. The models showed that these bacteria were expected to live harmoniously under conditions where expressing CAT had a fitness cost. When chloramphenicol resistance came at no expense, the CAT strain outcompeted the susceptible strain to create a homogenous population.
The team next infected mice with both CAT and susceptible bacteria or with only the susceptible bacteria. In the presence of antibiotics, the researchers recovered both bacterial strains under conditions that supported the fitness cost hypothesis.
When considering the wider implications of their findings, Sorg stated that bacteria other than Pneumococcus may be undertaking the worst sort of team work: “This could absolutely be happening with different antibiotics. It could even be occurring between species of bacteria, each carrying a different resistance gene and as a collective developing a multidrug resistance.”
Please enter your username and password below, if you are not yet a member of BioTechniques remember you can register for free.