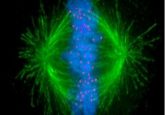
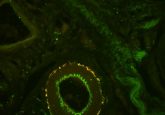
Blowing up brains
A new microscopy technique to expand tissue samples makes for a faster and more detailed way to image the whole fly brain.
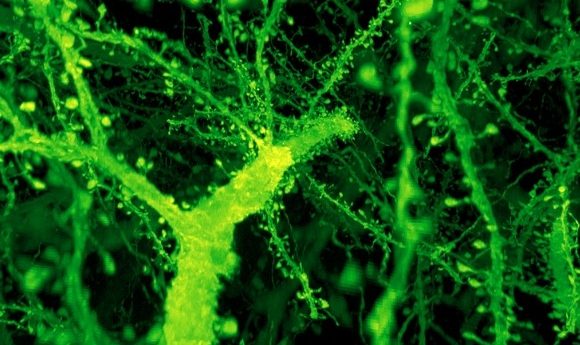
A forest of dendritic spines protrudes from the branches of neurons in the mouse cortex. Credit: Gao et al./ Science 2019
Ruixuan Gao and Shoh Asano from Ed Boyden’s lab at the Massachusetts Institute of Technology (MIT; MA, USA) have developed a new microscopy imaging technique that allows for the capture of the entire fruit fly brain in just 62.5 hours.
The technique, called expansion microscopy, involves using chemicals to make small specimens up to four-times bigger. By infusing tissue samples with a swellable gel, they found a way to expand the samples, locking the molecules in a gel scaffold that allowed them to maintain their relative positions while being less crowded and easier to see under a microscope.
The method had been shown to be effective on single cells or thin tissue sections, but the team wanted to image much larger tissue samples, allowing for the identification of complete neural circuits spanning millimeters or more.
Needing a microscope more advanced than that of their own lab, they turned to Eric Betzig and his research group at the Howard Hughes Medical Institute’s Janelia Research Campus (HHMI; VA, USA) for use of their lattice light-sheet microscope; a higher speed, higher resolution and gentler scope relative to what was previously available to them.
The lattice light-sheet microscope works by sweeping an ultrathin sheet of light through the specimen, illuminating only the section that is in the microscope’s plane of focus. This reduces the amount of light on the specimen and lowers the risk of photobleaching, helping to maintain the fluorescence of the rest of the tissue. “The result is that we get crystal clear images at blazingly fast speeds over very large volumes compared to earlier microscopy techniques,” explained Boyden.
Betzig was skeptical: “I thought they were full of it,” he remembered, though when the results came back positive, he was blown away. “I couldn’t believe the quality of the data I was seeing. You could have knocked me over with a feather.”
The images produced were detailed enough to show the small dendritic spines of neurons. The final data, recently published in Science, was the work of multiple groups working together to capture and analyze the images. “This is like an Avengers-level collaboration,” commented Gao.
Gao and Asano expanded and collected around 50,000 cubes of data for each brain, resulting in a kind of 3-D jigsaw puzzle. Computer scientists Stephan Saalfeld and Igor Pisarev, also of the HHMI, assisted in putting the images together. “They dealt with all the horrible little details of image processing and made it work on each multi-terabyte data set,” commented Betzig.
Co-first author Srigokul Upadhyayula (Harvard, MA, USA) analyzed the combined 200 terabytes of data and created movies, showcasing the brain’s intricacies in vivid color. In total the group investigated over 1,500 dendritic spines, counted all of the synapses in the brain, imaged myelin sheaths and highlighted each dopaminergic neuron.
The speed of this technique will allow scientists to answer questions such as how brain circuits vary and how brains differ between males and females with the end goal of mapping the entire nervous system. “We’ve crossed a threshold in imaging performance,” commented Boyden. “That’s why we’re so excited. We’re not just scanning incrementally more brain tissue, we’re scanning entire brains.”