Ruslan Dmitriev on FLIM and intestinal organoids


Ruslan Dmitriev
At ASCB 2019, BioTechniques caught up with Ruslan Dmitriev, group leader of the Metabolic Imaging Group at the University College Cork (Ireland) to discuss his work with fluorescence lifetime imaging microscopy (FLIM) and intestinal organoids.
Can you tell us about your presentation at ASCB?
I have given two presentations this year. I presented a poster on new biosensor scaffolds for tissue engineering and my second presentation was a microsymposium talk on FLIM, for imaging of the metabolism of intestinal organoids.
Can you tell us more about the techniques you use?
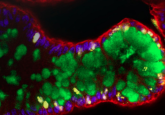
My research is centered around developing new high-performance biosensors, which enable you to do fluorescence lifetime imaging of various cellular processes such as metabolism, dynamics of lipid droplets, mitochondrial polarization and measuring oxygenation (hypoxia) in real time. The main model we use is adult stem cell-derived intestinal organoids, Lgr5-GFP. We try to make analysis multi-parametric, measuring at the same time with live state analysis of the stem cell niche and neighboring cells. We correlate how, during different physiological conditions (for example differentiation of the organoids growing a mini-gut), what’s happening to the cells and their metabolic needs, how they adapt and how this might affect their developmental fate. Another area very close to that is looking at the scaffold material in which the organoids grow. The use of Matrigel matrix for growing organoids is well accepted, and what we try to do is to make this Matrigel not only act as a supportive material, but to also display biosensor functions. In that case, you modify the Matrigel and can study what happens without cell labeling, for example, with oxygenation of the organoids. Potentially, you can also modify the cellular microenvironment to respond to such things as changes in pH so that the matrix can start remodeling and responding to the needs of the tissue that it surrounds.
If you would like to learn more about 3D models, check out our Spotlight on 3D cell cultures here!
When you’re using techniques like FLIM, are there any challenges that you come across and if so, how do you overcome them?
There are a lot of challenges. One is choosing the right biosensor and ensuring that the calibration is valid. Another is that we use custom-made microscopes and there can be a lack of standardization in approaches – other labs might do things in a slightly different way. We try to perform the analysis the same way as most other labs. Another challenge is choosing the correct software and having a data workflow that is more or less standardized, along with data analysis and statistics. However, the biggest challenge currently is to reconstruct FLIM images in 3D and a dynamic shape, and to do it relatively quickly, in a way that allows better understanding of what happens to the cells, with more precision from the data.
What techniques do you use to reconstruct those images?
We have laser scanning microscopy, and we also benefit from collaborators with 2-photon excited FLIM. We try to get the best available approaches in microscopy. We heavily rely on the use confocal microscopy to get sectioning perfect, and then reconstruct it in 3D using open-source software such as ImageJ.
- Organoids for the study of pancreatic cancer with David Tuveson
- Mini-brains created from primary cells
- Talking Tech News | Cerebral organoids: past, present, future
What challenges do you come across specific to intestinal organoids?
There are a lot of challenges. The major one is heterogeneity. When we started to grow organoids from adult stem cells we figured out that they come in very different sizes and shapes. We found that they depend on the selection of growth media – you buy a medium, for example, from vendor A and then you find that your organoids look a certain way, then you produce the medium by yourself and the organoids look very different (and sometimes more healthy). So, the setting up of the methodology can be challenging, although if you have good experience of working with other 3D models it might be a bit easier. The fact that organoids are three-dimensional is also a challenge. However, it can be quite exciting to overcome these challenges, and fun because they come in different sizes and shapes – sometimes you look under the microscope and they might have a shape of an animal or something else esthetically pleasant! The methodology can also be expensive, so another challenge is funding, but because we do microscopy imaging we don’t really need much of the material – you would need more money if you focus on western blotting or other kinds of next-generation sequencing analyses.
What do you love about investigating organoids?
The fact that it brings us a bit closer to the native tissue. People talk a lot about organoids now but intestinal organoids are one of the nicest examples, because you recreate all the complexity of intestinal epithelium, and it happens spontaneously and in a self-organized manner, so basically the cells need very little in terms of environmental cues to shape them. It also gets you thinking about the 3Rs, the reduce, refine, replace strategy to minimize animal experimentation. That’s very exciting.
Is there any cell information that we currently can’t access with the imaging techniques that we have today?
Yes absolutely, there are a lot of things to discover! You might think you’ve discovered everything, but then a new methodology appears and you get a new set of data and you have to build theories around that. There are a lot of exciting things in the viscosity, the membrane composition and physical parameters that people normally can’t access and hopefully a FLIM can be useful for that. Also, how you can trace different cell types? There are a lot of new cell types that have been discovered within the intestine just thanks to the organoid methodology. I hope with the introduction of FLIM to organoid research you will be able to trace more accurately and distinguish cell types within the stem cell niche based on their metabolic needs and how they are interconnected.
- Anne Carpenter on artificial intelligence in the cell imaging field
- Nano-sized laser: the future for imaging living tissues?
- Artificial intelligence set to revolutionize cell imaging methods
What would you need to change or improve in current imaging techniques to enable you to uncover more of that information?
The main bottleneck at the moment is that the methodology is mostly attractive for specialists – photo-optical instrumentation engineers and ‘photonics’ people – the people with a physical instrumentation background that enables them to interpret their result. However, I think that a number of vendors already make it more user-friendly. Streamlining data processing and speed are the challenges that have to be overcome. What’s more, with the production of big data, a challenge is the introduction of more bioinformatics approaches in order to interpret the data and to analyze them.
Where do you see the field of live cell imaging going in the next 5 years?
I may be biased, but first I see the wider introduction of biosensor scaffolds and also the engineered scaffold materials that will respond dynamically to the growth of the developing tissue, for example changing the softness or permeability of gases during the different stages of embryonic development. I definitely see the introduction of more multi-parametric assays; FLIM is one of the methods as well as Raman – especially non-destructive coherent anti-Stokes Raman spectroscopy (CARS). There are a lot of label-free methodologies appearing at the moment and they provide you with single-cell subcellular resolution without any dyes. They show very good promise. I also hope the area will move more towards being quantitative, multi-parametric and live, and overall a relatively non-invasive assessment of life at the single-cell or subcellular levels.
Do you have any advice for best practice when you’re constructing scaffolds?
Try to get to the people who are working next door; be interdisciplinary. For example, if you’re a stem cell biologist try to collaborate with people from tissue-engineering areas, biomedical materials engineers and bio-informaticians.
When you were starting out in your career, did you have a particular hero who inspired you to go down your career path or was it more of an organic process for you?
You always have some people that inspire you, but for me one of the inspirations has been going to meetings like the ASCB Meeting where you can have a chance to talk and listen to amazing keynote speakers, and even though a talk might be outside of your area, you can really see what else you can do in your research problem and how far your ambition can go.
In a more specific sense, I think, someone who continues to inspire me is Leonardo da Vinci – a perfect artist and scientist at the same time.
My PhD supervisor back in Russia was also very important for my choice to become a scientist – he inspired me in the ways he behaved and approached research problems.