Culturing cells from patient brains
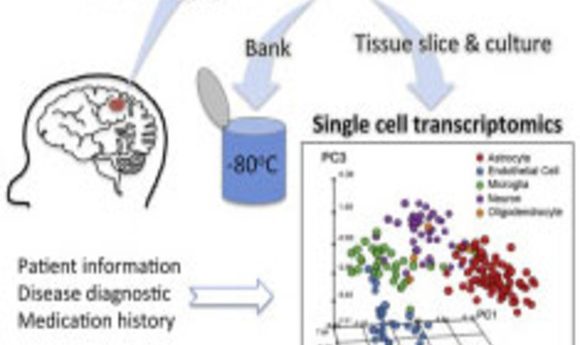
Researchers performed single-cell transcriptomics on cultured live-adult human brain cells, furthering the goals of personalized medicine and opening a door for understanding human cognition.
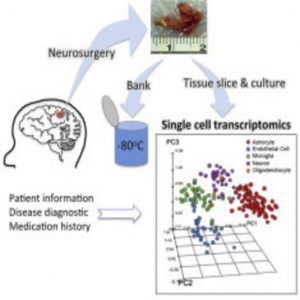
Diseases and drugs affect various cell types in different ways, and investigations into these effects have been limited by the lack of a system for single-cell analysis of live-patient-derived brain cells. For the most part, single-cell neuron work has been performed in rodents, while cell-type studies in humans have been largely limited to postmortem studies, cancer cell lines, and more recently, acute harvest of cells from patients. Although these studies provide valuable human transcriptomic information, the acute harvest of cells does not allow for morphological or long-term functional investigations, other than sequencing.
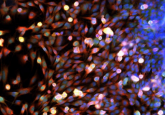
Recently, Junhyong Kim, co-director of the Penn Program in Single Cell Biology at the University of Pennsylvania, and his colleagues reported in Cell Reports that they had developed a culture system for healthy adult human brain cells from patient biopsies collected at the time of surgery. Using these cultures, they identified transcriptional hierarchies specific to individual cell types and patients. According to the authors, single-cell transcriptomics on cultured live-patient-derived cells is a prime example of the promise of personalized precision medicine.
“This particular study involved some of the first human brain cells analyzed from living patients,” said Kim. “Understanding the unique molecular characteristics of individual cells will help us understand the mechanisms of human diseases like Alzheimer’s, autism, and Parkinson’s, and ultimately, how individual cell function contributes to human cognition.”
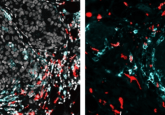
Kim’s team cultured cells from the biopsies for up to 84 days and then performed deep sequencing of hundreds of single cells to obtain their individual RNA expression profiles, allowing them to capture the range of expression for each cell type.
By performing single-cell transcriptomics on more than 300 cells, Kim and his collaborators identified specific cell types, such as neurons and astrocytes, after 3 weeks in culture. Using deep sequencing, they detected more than 12,000 expressed genes, including hundreds of cell type–enriched transcripts (e.g., mRNAs, long non-coding RNAs, and primary miRNAs).
Because these cells derived from subjects ranging in age into their sixties, this system permits human aging studies previously possible only in rodent systems. In addition to providing a model system to study human disease and drug treatments, the findings provide insight into the proper functioning of the human brain.
According to Kim, the next challenge will be to make more precise and robust measurements while scaling up to high-throughput processing techniques to assay millions to billions of cells in an organ. “We showed the feasibility of obtaining single-cell data from live human brains,” he said. “The next step will be to ramp up our work to sufficient scale to address the entire brain.”