DNA replication in a traffic jam
What happens to DNA replication when its components compete with other processes for access to the genome?
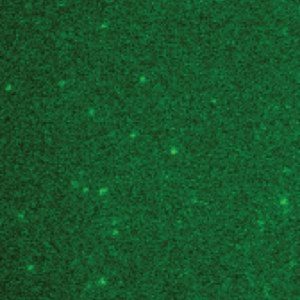
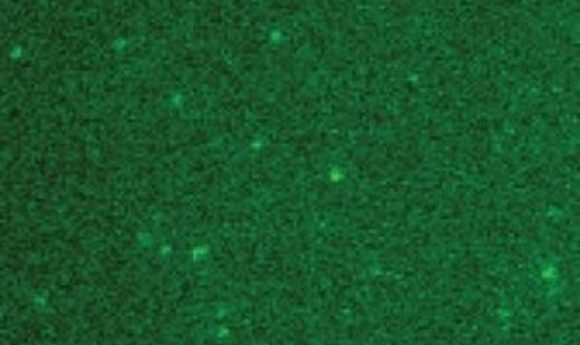
Purified surface immobilized single GFP molecules on a slide (1).
DNA replication is often depicted as an orderly linear process, where replication proteins and enzymes (the replisome) assemble at the origin. However, chromosomes are very busy places. So what happens when the replisome runs into a transcription complex?
The research groups of Houra Merrikh and Paul Wiggins at the University of Washington recently addressed this question. Writing in eLife, they build on previous biochemical and genetic studies to examine replisome kinetics in living Escherichia coli and Bacillus subtilis, using single molecule fluorescence microscopy (SMFM).
The group followed a GFP-labeled replisome component (DnaC) in these species and found that while there was only one active replisome complex per cell, the complexes had relatively short lives. Historically, the replisome has been expected to be very stable, completing replication without many interruptions. Our results, however, indicate that contrary to the accepted view, the replisome collapses and the machinery disassembles multiple times per cell cycle.” Merrikh noted.
To examine the possible causes of replisome disassembly, the researchers treated the bacteria with a transcription inhibitor and observed DnaC dynamics in rpoB mutants, which have an unstable RNA polymerase. In both cases, the replisome complexes persisted much longer than controls, showing that gene transcription directly interferes with DNA replication. They also observed that the replisome collapses when it encounters a highly induced reporter gene in an orientation opposite to the direction of replication.
This approach, while powerful, presented some challenges. “Visualizing single molecules is very difficult because the low levels of fluorescence intensity make it difficult to separate the signal from noise. Counting the number of DNA-bound replisome proteins is particularly challenging because it requires us to detect the decrease in fluorescence intensity associated with a single molecule.” Merrikh said.
Despite these challenges, Merrikh’s team shows that DNA replication in bacteria is not a smooth continuous process due to the instability of the replisome as it collides with transcription machinery. Merrikh added, “Other obstacles to the replisome include DNA-binding proteins and breaks or lesions on the DNA itself. We have not directly observed the fate of the replisome in the face of these obstacles, but they could certainly cause disruption.”
While the group studied replisome dynamics in bacteria, Merrikh said that their findings could apply to more complex organisms. “There is biochemical evidence that replication-transcription conflicts also occur in eukaryotic cells, and the replication proteins themselves are highly conserved between organisms.”