Medicated microbiomes
New research reveals that individual variation in gut microbiomes might make medications ineffective for some.
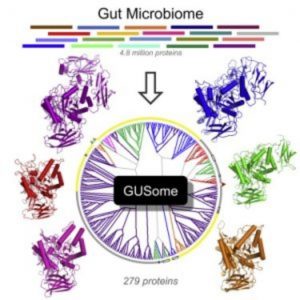
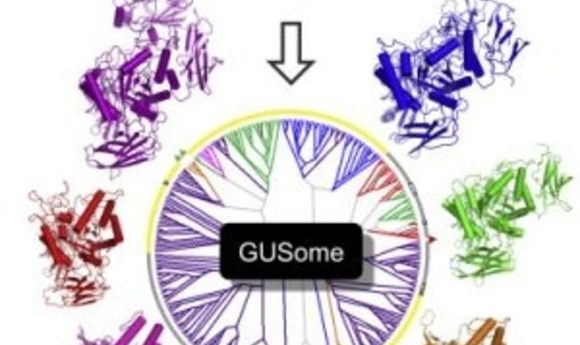
An atlas of β-Glucuronidase (GUS) enzymes from the Human Microbiome Project (3).
Medications and treatment plans are designed based on averages, but statistically we can’t all be average; a treatment that works for one might not work for another. In the new wave of personalized medicine, clinicians are tailoring treatment plans with the hope of helping those whose response is outside the average range. Thus far, personalized diagnosis and pharmacological interventions for disease have largely focused on genomics. However, increasing evidence suggests that the microbes hanging out in the human gut also play an important role in drug response.
“People have known for a long time that the microbiome can interact with drugs, but it’s only recently that we’ve really been able to sort of target the specific microbial culprits,” said Libusha Kelly from the Albert Einstein College of Medicine.
Thinking about the microbiome might bring to mind the digestion and metabolism of food. However, the microbiome is remarkably plastic and adapts to shifts in the environment. In addition to the turkey sandwich someone eats for lunch, the bacteria in their gut will also gobble up any medications, which may cause problems for drug efficacy depending on the makeup of the unique microbiome.
“Imagine that you have a drug like sulfasalazine, which is used for rheumatoid arthritis and irritable bowel disease,” said Kelly. Sulfasalazine is activated in the gut by the enzyme azoreductase. As variability exists in the gut microbiomes in the population, some patients make less of the enzyme. “That might suggest that you’re getting less of the active form of the drug than you actually want.”
Going With the Gut
Although researchers design each drug to directly treat a problem, off-target activities sometimes cause pronounced side effects that ultimately limit the drug’s efficacy. This is the case with irinotecan, a commonly used chemotherapeutic agent that becomes problematic when, during digestion, it undergoes a second round of activation in the gut by β-glucuronidase enzymes, leading to severe diarrhea. If researchers could inhibit the reactivation of the drug, could they also eliminate the undesirable side effects?
In a 2010 paper published in the journal Science, Matthew Redinbo and his colleagues at the University of North Carolina at Chapel Hill did just that [1]. Using high-throughput screening, the researchers identified compounds that effectively inhibited the β-glucuronidase enzyme, but did not disrupt the commensal bacteria necessary for basic human health. To test the efficacy of inhibitors in vivo, the researchers administered them alongside irinotecan and found a significant reduction in drug-induced toxicity. “The non-lethal modulation of microbiome targets using small molecule inhibitors is a viable approach to working with gut microbes, not against them as antibiotics do,” said Redinbo. These findings demonstrate that it is possible to alleviate the negative side effects of a drug in order to improve tolerability and efficacy.
Kelly first came across Redinbo’s paper while a postdoctoral fellow at the Massachusetts Institute of Technology. Fascinated, she recently decided to further investigate this question as she started her own lab at the Albert Einstein College of Medicine.
Moving away from animal models, Kelly and her colleagues collected fecal samples from 20 healthy individuals in order to examine how unique microbiomes affected irinotecan metabolism. When the researchers treated the fecal samples with irinotecan and collected and analyzed the compounds produced by the bacteria, they found that a fifth of the samples produced the toxic form of the drug.
Interestingly, further analysis revealed that samples from these high-metabolizers housed strains of bacteria that contained more β-glucuronidase, the pesky enzyme known to reactivate the drug as a toxic compound. Although this collection of 20 healthy individuals was not meant to represent cancer patients actively taking irinotecan, the study is one step toward a better understanding of how individual variation in the microbiome can render therapies ineffective for some.
A Spoonful of Enzymes Helps the Medicine Go Down
Kelly and her colleagues would now like to conduct a similar experiment in cancer patients taking irinotecan. “The thing that I hope for most in the clinic is that we can use non-invasive fecal samples to predict—before we give a person a drug—how they’re likely to metabolize it,” said Kelly.
These studies may open the door for additional clinical trials combining currently available cancer therapies with enzyme inhibitors to offset the negative side effects. However, our understanding of the numerous enzymes at play in gut-drug interactions is still very limited. For example, although inhibiting an enzyme might improve a drug’s side-effect profile, the enzyme may be involved in other activities necessary for maintaining a healthy body.
Studies like these leave researchers wondering if there are more targeted ways to interact with the microbiome that would make people less likely to have adverse responses to a drug. Furthermore, it’s still unknown how much microbiome variability actually exists within the human population. It may take some time before clinicians understand bacteria-drug interactions well enough to more effectively treat a patient in a personalized therapy setting, but these studies are a step in the right direction.
Earlier this year Redinbo and colleagues published the first comprehensive atlas of drug targets in the human microbiome [3]. Before scientists can begin targeting specific enzymes they must first know which enzymes are present and how they function in the gut. “The field is moving impressively fast toward mechanistic connections between specific microbial factors and their effects on mammalian health,” said Redinbo. “Fortunately, we can used the well-established tools of structural biology and biochemistry to understand individual microbiome enzymes and then begin to place them in context within microbiota communities and symbiosis with mammalian systems.”