Playing with genetic alphabets
Four nucleotide bases carry all the information an organism needs to live and function. Can this limited set of bases be expanded to carry more information?
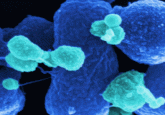
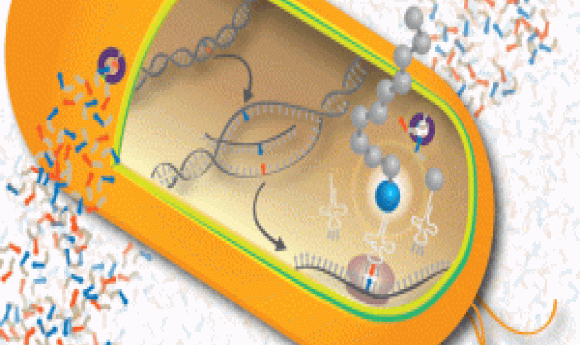
Unnatural bases shown in red and blue undergoing DNA replication, transcription, and translation to create a new protein with an unnatural amino acid (blue ball). The nucleic acid transporter for unnatural bases is shown in purple.
Image credit: Romesberg Lab
The central dogma of molecular biology is that DNA is transcribed into mRNA, which is then translated into proteins. The whole process relies on hydrogen base pairing of the nucleotides A, C, G, and T. Synthetic biologists recently began manipulating the gene expression machinery to create novel proteins and expand the functions an organism can carry out. Now, a new study by Floyd Romesberg at The Scripps Research Institute reports that introducing unnatural nucleotides into DNA results in the incorporation of novel amino acids into proteins in E.coli.
While other research groups have engineered cells to incorporate unnatural amino acids into cellular proteins, Romesberg focused on expanding the information stored in DNAso that semi-synthetic organisms could create new proteins and perform unnatural functions. “The natural system was there, and I wanted to augment the natural system,” said Romesberg.
The challenging journey began in 1999 when Romesberg established his own lab and explored hydrophobic base pairing for new bases as an alternative to complementary hydrogen base pairing with natural bases. He and his team set out to generate hydrophobic nucleotide analogues to eliminate the possibility that the naturally occurring bases would pair with them. “We used a medicinal chemistry approach where we made more than 200 different nucleotides and analyzed them in a test tube,” he recalled. “Initially they were so bad—we couldn’t do PCR and we had to force polymerases to take them in.”
Over time, they improved their chemistry and the analogues became polymerase compatible allowing DNA replication occur. By 2013, they had their first base pair of hydrophobic bases, which they tested in E.coli. “We were very surprised when it worked!” said Romesberg. “This emphasizes how important the 15 years of chemistry optimization was.”
With further optimization, the team made bases that replicated, transcribed, and translated into a semi-synthetic protein. “People want to me to say that it grew an eyeball or third arm but what is important is that the cells and the proteins didn’t do anything different,” said Romesberg.
Romesberg believes that the real impact of this work lies in the endless possibilities for functionalizing proteins such as tagging naturally occurring proteins for easier detection. In the future, they hope to move forward working with more complex organisms such as eukaryotic cells to see if they can validate the same phenomena.