RNAi: dust mites do it differently
RNAi plays a critical role in maintaining genome integrity in most multicellular organisms—but not in dust mites, a new study shows. What does this tell us about parasite evolution?
Dust mites (Dermatophagoides farina) are not parasites, but they descended from a parasitic species. This switch from parasitic to free-living form required significant genomic rearrangements that may involve transposable elements (TEs): long, repetitive DNA sequences that can amplify themselves and insert randomly throughout the genome.
TEs can wreak havoc on the genome, resulting in disease or sterility. Thus, organisms have adapted ways to silence them that include RNA interference (RNAi). Mosharoff Mondal and Alex Flynt at the University of Southern Mississippi, and Pavel Klimov at the University of Michigan examined RNAi pathways in dust mites and found they lack the conventional piwi-associated (piRNA) pathway used by most multicellular organisms to control TEs. Their findings could provide insight into how parasites evolve.
“Its very strange for an arthropod to have RNA-directed RNA polymerase (RDRP),” noted Flynt, lead author of the study. “We were looking for an organism that had RDRP and found that dust mites have it, but they do not have piRNA, which was also very strange.”
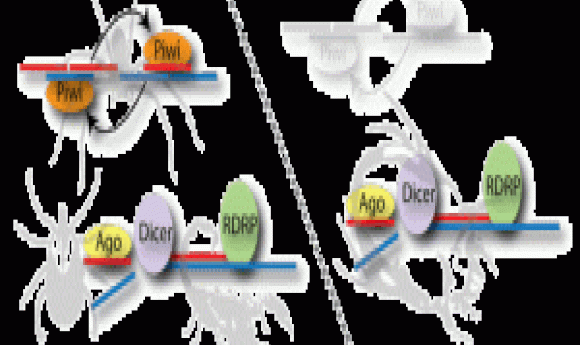
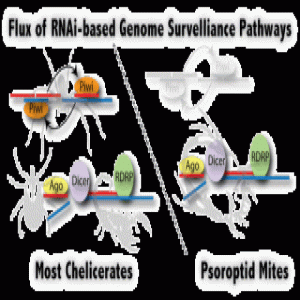
piRNA and RDRP-mediated RNAi are used in two mechanistically different RNAi pathways. Arthropods and vertebrates use the piRNA pathway or “ping-pong” mechanism, where small RNAs target TE transcripts to form a hybrid RNA complex that is cleaved by Zuc nuclease and amplified by passing between Zuc and Piwi proteins; the piRNAs generated by this process silence TE transcription. In contrast, RDRPs occur in plants, fungi, and nematodes. In these phyla, RDRP converts the TE transcript to a dsRNA that dicer nuclease cleaves into siRNAs (small-interfering RNAs). Argonaute proteins then target these siRNAs to TEs in the genome and silence their expression by modifying chromatin.
To identify which pathway dust mites use, Flynt and his colleagues sequenced the dust mite genome. They found that dust mites possessed genes coding for argonaute proteins, suggesting an RDRP pathway. They then sequenced dust mite small RNAs, analyzed dust mite RNA size distribution, and compared these with small RNAs from spider mites, which use the piRNA pathway. The dust mite small RNA size distribution and sequence was consistent with an RDRP pathway. They validated the RDRP model by silencing dust mite dicer genes in vivo and observed a significant increase in TE expression.
Flynt believes that the absence of the piRNA pathway in dust mites can offer insight into its evolutionary history and how organisms become parasitic. “The dust mite evolved from a parasite related to the scabies mite. It is really interesting to consider why an organism became a parasite because there has to be genetic mayhem for it to make the leap to a host-dependent lifestyle. This has also been seen in parasitic flatworms, which have also lost the piRNA pathway. So loss of piRNA might be a way to get massive genome rearrangements and acquire the parasitic form,” he said.





