Splitting the details of protein aggregation
Two new tools for quantifying protein aggregation promise to amplify our understanding of prion function and inheritance and the role of protein aggregates in healthy and diseased cells.
What is infectious but not alive, transfers across generations, but is not genetic material? Currently, the only answer to this riddle is a prion.
Prion proteins fold into complex quinary structures that can aggregate to disrupt cellular function. They also travel through the cytoplasm to enter other cells lacking prions. In mammals, prions are notorious for their roles in neurodegenerative diseases such as mad cow disease, Parkinson’s disease, diabetes, and Alzheimer’s disease. However, in some cases, they are also involved in regulatory and beneficial cellular functions. For instance, yeast prions serve in functions related to their survival, evolution, and metabolism.
Investigating prion formation and its influence on function has gained prime importance in recent years. However, until now, protein aggregation studies relied upon investigating consequent phenotypic changes, structural analyses of certain types of aggregates, or biochemical assays on lysed cells. None of the methods commonly used for those studies is suited for quantitative, in vivo, and high throughput analysis, which is the biggest need right now, according to Ahmad Khalil from Boston University.
One Tool Fits All
Inspired to solve this problem, Khalil led a team of researchers in designing a quantitative, generalized tool for tracking protein aggregation in vivo that was not limited to studying one specific protein [1]. At the onset of the study, one of his main goals was to develop a tool that functions across many different types of structures and in many cellular contexts, returning a simple readout amenable to screening.
His team came up with a clever strategy; they fused a protein of interest with a sensitive synthetic transcription factor that activated a reporter gene to express a fluorescent protein. The state of the protein of interest modulated the transcription factor function; in its soluble state, the protein activated the reporter, yielding a fluorescent readout as the cells passed through a flow cytometer, while in its aggregated state, it failed to activate the reporter gene, thereby producing no fluorescence.
“The strength of the design is the synthetic transcriptor, which allows it to tune this system to any protein of interest,” said Khalil.
After the researchers verified that their tool, yTRAP, could successfully monitor protein aggregation in yeast, they developed several variants of the design. The reverse yTRAP tool, for instance, possessed a positive reporter system using a transcriptional repressor instead of an activator so that protein aggregation turned on the expression of a fluorescent reporter protein. Other variants included a dual sensor to monitor multiple prion protein states in cells or a sensor that identifies mutations that eliminate prions from the progeny cells for anti-prion drives.
“This is a really useful tool that at least demonstrated in yeast that it can enable quantitative, high throughput studies on the aggregation phenomenon,” said Khalil.
Yeast prions can also be adapted for use as synthetic cellular tools for encoding memories of an environmental trigger on protein expression in yeast progeny or for studying associated phenotypic changes, according to Gregory Newby from Massachusetts Institute of Technology, first author of the study. “High throughput screening with yTRAP can be used to extend these tools to identify new prions and prion-switching factors, building this toolbox of synthetic prion components,” he added.
Single Flurophore, Dual Fluorescence
Concurrently, Randal Halfmann from the Stowers Institute for Medical Research independently worked on developing a tool to probe prion formation [2]. Since prions are formed probabilistically, meaning that the system will stabilize over time to form these structures provided that the protein concentration crosses a critical threshold, Halfmann wondered if nature exploits nucleation barriers, influenced by protein concentration and time, to regulate protein function in the cell.
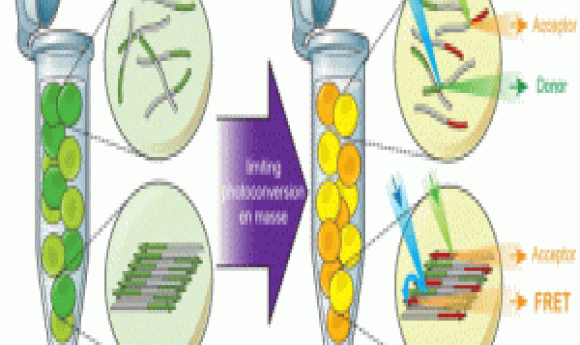
To understand the influence of protein density arising from spatial constraints on prion formation, Halfmann’s team needed a tool that reported protein assembly in a concentration dependent manner. The researchers were inclined to incorporate fluorescent reporters for direct visualization, but wanted to avoid fusing dual flurophores to the protein to avoid challenges in maintaining a fixed flurophore ratio across several cells, an important criteria for scaling single cell data to the entire cell population.
Halfmann’s team chose a unique approach that satisfied all their conditions and developed Distributed Amphifluoric Fluorescence Resonance Energy Transfer FRET (DAmFRET), which consisted of the protein of interest fused to a single green fluorescent protein construct that converts to a red florescent form when stimulated with violet light. By controlling the dosage of illumination, the team achieved an optimum ratio of green:red molecules for maximum FRET efficiency. When proteins aggregated, the researchers saw increased red signals resulting from FRET between flurophores on adjacent proteins. They detected these changes using flow cytometry.
“DAmFRET helps us quantify the nucleation barrier that helps us determine if a protein is a prion, how frequently it will form a self assembled prion state, and how long an organism will have to wait until it happens,” explained Halfmann.
In this study, the team applied their tool for screening signaling proteins and found that proteins involved in the innate immune response functioned via prion like switches. In the future, Halfmann’s team plans to study the consequences of nucleation barriers apart from function. “Any supersaturated protein will inevitably acquire the nucleated state if the system waits long enough,” he said. This means that the probabilistic nature of nucleation provides a molecular mechanism for ageing. “We are most excited about that aspect,” he added.
Although both yTRAP and DAmFRET involved flow cytometry, Halfmann pointed out that DAmFRET is more quantitative than yTRAP. According to him, although yTRAP distinguished between cells with or without prions, it does not provide information regarding the concentrations required to form aggregates.
Khalil noted that yTRAP is susceptible to false positives and negatives since the transcription factors may be modified by other mechanisms, but he emphasized that, “Just like other techniques, it provides high power and capability.”
Khalil’s team now plans to extend this tool to other cell types. “It is designed to be synthetic and agnostic to cell type, so we think we will be successful there,” he said.
“As with most other insights gained from yeast, it is likely that these discoveries will directly speak to the biology of other eukaryotes, including humans,” added Newby.