Time to understand the oscillator in a circadian clock
Structural studies revealed the role of protein interactions in cyanobacterial circadian rhythms. What does this tell us about biological clocks in complex organisms?
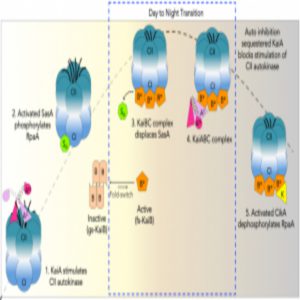
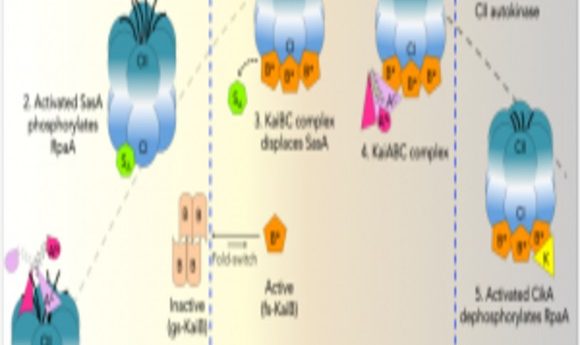
Model of rhythmic interactions of protein complexes involved in the cyanobacterial clock.
Credit: Archana Chavan, LiWang lab, University of California, Merced
Most people experience jet lag after traveling long distances as their body becomes confused about the altered eating and sleeping times. This is because all organisms, from bacteria to humans, possess an in-built internal clock to cope with the daily light and dark cycles. Thus, even if the external clock timing changes during travel, the internal body clock that governs metabolism needs time to adjust.
Researchers have long known about the existence of circadian rhythms, but the detailed underlying mechanism for how it works remains hazy. Recently, researchers from the University of California, Merced determined how periodic conformational changes in the circadian protein complexes in cyanobacteria influence gene regulation. Archana Chavan, lead researcher of this study, presented her team’s findings last month at the Biophysical Society meeting in San Francisco, California [1, 2].
“The fundamental mechanism of biological clocks remains the same in all living organisms,” said Chavan. Therefore, she chose the easy-to-cultivate cyanobacteria as a model organism. The cyanobacterial circadian clock is unique; it is the only clock wherein all proteins involved in the time-keeping mechanism can be isolated and tested in vitro.
In her experiments, Chavan analyzed the structural interactions of the three main proteins that together constitute the circadian clock oscillator in cyanobacteria, KaiA, KaiB, and KaiC, in the presence of ATP over 24-hour periods using nuclear magnetic resonance (NMR). “We put our sample in this big magnet and it generates selective frequency (rf pulse), and using different frequencies, we can actually look at particular atoms in the molecule,” explained Chavan.
The team noted cyclic binding-unbinding of the Kai family proteins, wherein the master regulator KaiC bound KaiA during the day and KaiB during the night. These interactions triggered a signaling cascade, thus regulating specific downstream genes; in the daytime, genes responsible for metabolizing sugars produced by photosynthesis were expressed, while genes driving cell division were expressed at night.
“This is indeed a very nice piece of work by the LiWang lab. Following the temporal behavior of protein complexes by NMR beautifully illustrates the power of the technique when applied to a system where time plays a pivotal role,” said Angela Gronenborn from the University of Pittsburgh School of Medicine, who was not involved in this study.
Chavan believes that understanding the basic principles of circadian clocks in simplistic bacterial models will help unlock the mysteries of biological clocks in complex organisms in the future.