No, we don’t have a ‘lizard brain’

Despite reptiles and mammals having gone through hundreds of millions of years of separate evolution, researchers identify a core set of neuron types present in both brains.
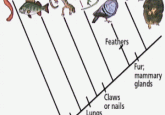
A mere 320 million years ago, early tetrapods ventured out of the water and onto land, marking a turning point in vertebrate evolution. This move gave rise to reptiles, birds and mammals, which are three of the major clades, groups of organisms that share a common ancestor, of life that escaped the sea.
In the 1960s, Paul MacLean (Yale University, CT, USA) proposed the idea of the triune brain based on anatomical studies, suggesting vertebrate brains are made of a core ‘lizard brain’ with the addition of specific features that evolved in stages after life moved to land. However, researchers at the Max Planck Institute for Brain Research (Frankfurt, Germany) have revealed that most areas of mammalian and reptilian brains have a mixture of common neuron types, from their ancient ancestors, and clade-specific neuron types.
This finding finally puts the idea of a ‘lizard brain’ to rest. Gilles Laurent, who led the study, explained: “Distinct brain areas do not work in isolation, suggesting that the evolution of interconnected regions, such as the thalamus and cerebral cortex, might in some way be correlated. Also, a brain area in reptiles and mammals that derived from a common ancestral structure might have evolved in such a way that it remains ancestral in one clade today, while it is ‘modern’ in another. Conversely, it could be that both clades now contain a mix of common (ancient) and specific (novel) neuron types. These are the sorts of questions that our experiments tried to address.”
Shapeshift, eat brains, disclose evolutionary secrets: a day in the life of an amoeba
A unicellular organism that can rebuild itself into a different organism, may also be a crawling, swimming archive of evolutionary knowledge. When it’s not eating your brain.
Instead of relying on anatomical differences, the research team created a molecular atlas from the brain of an Australian bearded dragon Pogona vitticeps using single-cell RNA sequencing and compared it to existing mouse brain datasets. Single-cell RNA sequencing has a higher resolution than traditional approaches used for comparing brain regions and was able to expose similarities and differences. They profiled over 280,000 cells from the Pogona vitticeps brain and identified 233 distinct neuron types.
“Computational integration of our data with mouse data revealed that these neurons can be grouped transcriptionally in common families, that probably represent ancestral neuron types,” explained David Hain, the co-first author of the study.
Their analysis discovered that reptilian and mammalian brains evolved clade-specific neuron types and circuits from common sets of ancestral features. They also found a core set of neuron types that possess similar transcriptomes, which aren’t restricted to one ‘lizard’ region of the brain.
Additionally, co-first author Tatiana Gallego-Flores implemented histological techniques to map these cell types in the brain. In doing so, they observed that neurons in the thalamus could be grouped into transcriptomic and anatomical domains, which are defined by how the neurons connect to other regions of the brain. These regions have different outcomes in mammals and reptiles, with either conserved or divergent neuron types. Comparing the transcriptomes of the two domains revealed that transcriptomic divergence corroborated with the target regions.
“This suggests that neuronal transcriptomic identity somewhat reflects, at least in part, the long-range connectivity of a region to its targets,” concluded Laurent. “Since we do not have the brains of the ancient vertebrates, reconstructing the evolution of the brain over the past half billion years will require connecting together very complex molecular, developmental, anatomical and functional data in a way that is self-consistent. We live in very exciting times, because this is becoming possible.”