Novel microscopy technique provides insights into amyloid architecture
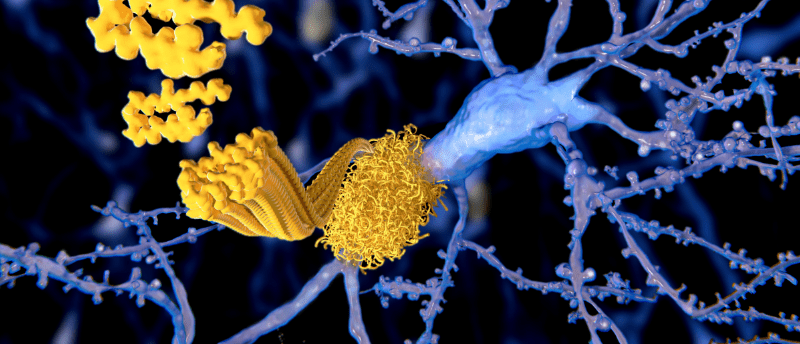
Researchers have developed a microscopy technique to visualize amyloid beta’s underlying structure, which could yield new insights into Alzheimer’s disease.
As the prevalence of Alzheimer’s rises, understanding the underlying pathophysiology of the disease is crucial to finding new treatments. Now, researchers from Washington University in St. Louis (MO, USA) have developed a new microscopy technique to visualize how amyloid beta (Aβ) structures, which are implicated in Alzheimer’s, grow and decay.
Aβ is the main component of amyloid plaques, the deposits found in the brains of individuals with Alzheimer’s disease. Aβ peptides aggregate into different types of structures, including oligomers, protofibrils and amyloid fibrils, and it’s these amyloid fibrils, which are larger and insoluble, that further assemble into amyloid plaques [1]. However, current techniques have struggled to determine how Aβ structures grow and decay, only showing final structures and not how they’re organized.
“The individual proteins are always changing in response to their environment,” said corresponding author Matthew Lew. “It is like having certain Lego bricks causing other bricks to change their shape. The changing architecture of the proteins and the assembled aggregates together leads to the complexity of neurogenerative disease.”
 Researchers discover new ALS gene
Researchers discover new ALS gene
Researchers have found a new disease-causing mutation in people with amyotrophic lateral sclerosis in a small region of Spain.
The team developed a new technique called single-molecule orientation–localization microscopy (SMOLM), which uses the flashes of light from chemical probes to visualize the sheets of peptides underlying Aβ42 fibrils, one kind of Aβ peptide [2]. SMOLM allows the researchers to look at the individual orientation of the underlying beta-sheets to see the relationship between their organization and overall structure.
Using SMOLM, the researchers found that, as expected, stable Aβ42 fibrils tend to retain stable underlying beta-sheets. The underlying beta-sheets of growing fibrils become more rigid and defined as they grow, whereas beta-sheets of decaying fibrils become increasingly less rigid and disordered.
The team also discovered that Aβ42 can grow and decay in unexpected ways. For example, sometimes fibrils can grow by peptides just accumulating, without the underlying beta-sheet orientations changing. Similarly, Aβ42 can decay as peptides leave, while the beta-sheet structure remains unchanged. They also found that Aβ42’s beta sheets can reorganize and change orientations without any immediate changes to the overall shape, but which may predispose to future large-scale remodeling.
“Because SMOLM can track Aβ42’s underlying organization and not just its shape, we can see different kinds of subtypes of remodeling that aren’t visible to diffraction-limited, non-orientation imaging modalities,” commented first author Brian Sun.
Determining how amyloid structures grow and decay will help researchers better understand amyloid’s role in neurodegenerative diseases like Alzheimer’s and could potentially aid in the development of much-needed therapies.