Harnessing the natural G=genome-editing capability of marine microbes
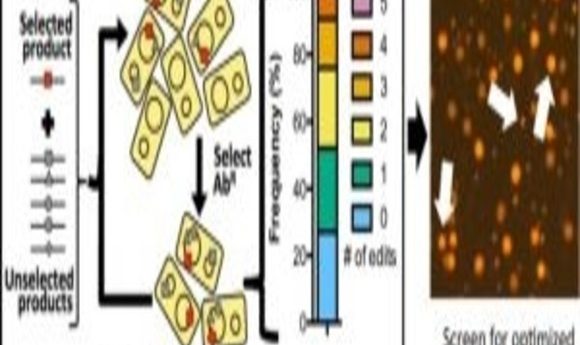
A new technique called MuGENT promotes the integration of multiple genome edits at high-efficiency on unprecedented timescales.
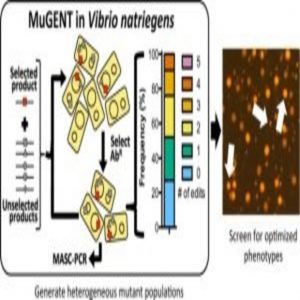
As the fastest growing organism known to mankind, the marine bacterium Vibrio natriegens has the potential to be exploited for diverse molecular biology and biotechnology applications. Now, in a study published in ACS Synthetic Biology, researchers report the development of a simple technique for rapidly generating highly complex mutant populations of V. natriegens.
According to the authors, their method for multiplex genome editing by natural transformation (MuGENT) will enhance the prospects of using this species for a variety of academic and industrial applications. “We think this will be a foundational method that will transform the way people work with V. natriegens,” said senior study author Ankur Dalia from Indiana University.
With its rapid growth rate (a doubling time of less than 10 minutes), broad metabolic capabilities, and lack of pathogenicity, V. natriegens is currently being explored as an attractive alternative to Escherichia coli. But low-efficiency genetic tools have limited the exploitation of V. natriegens for molecular biology and biotechnology applications. Although genetic techniques have been developed for V. natriegens, they are relatively laborious, require multiple steps, and must be used sequentially to generate multiple genome edits.
To address this problem, Dalia and his team turned to relatives of V. natriegens for a potential solution. Certain Vibrio species can take up DNA from the environment and integrate it into their genome by homologous recombination through processes known as natural competence and natural transformation, respectively. In the new study, the researchers discovered that V. natriegens is also capable of natural competence and high rates of natural transformation, with very little DNA required.
Next, the researchers tested whether they could harness natural transformation to simultaneously introduce multiple genome edits in a single step. When they used MuGENT to target 5 genes, they found that approximately 70% of the population contained at least 1 genome edit, and approximately one-quarter of the population contained 3 or 4 genome edits. The findings demonstrated that MuGENT rapidly and efficiently generated V. natriegens strains with multiple large genome edits without off-target effects, and it can be used to make highly complex mutant populations.
In additional experiments, the researchers demonstrated the utility of MuGENT for metabolic engineering by targeting the expression of nine genes involved in the biosynthesis of a chemical called poly-β-hydroxybutyrate (PHB). Following 4 cycles of MuGENT, which took just 5 days to perform, approximately half of the population had 3 or more genome edits, and approximately 10% contained 5 or more genome edits. Within 1 week, the researchers isolated edited strains that produced approximately 100 times more PHB than the parent and more than 3 times the amount of PHB than a rationally designed strain with just a single mutation. “This would not be possible using the classical genetic approaches that were available prior to this study,” Dalia said.
Unlike other multiplex editing approaches, MuGENT requires no specialized equipment and therefore has the potential to become commonplace. “One weakness of the MuGENT method outlined in this study is that making the synthetic DNA that will be used to manipulate these bacteria can be time-consuming,” Dalia said.
Moving forward, Dalia and his team will continue to develop and improve the methods for manipulating V. natriegens and start to develop this species for novel biotechnology applications. They are also interested in using these genetic tools to dissect the biology of this organism to better understand the mechanisms that allow for its remarkable growth rate.