Revealing thousands of rare cancer-causing mutations
By focusing on protein families instead of single genes, researchers found thousands of rare genetic mutations that influence cancer growth.
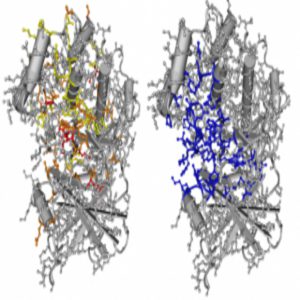
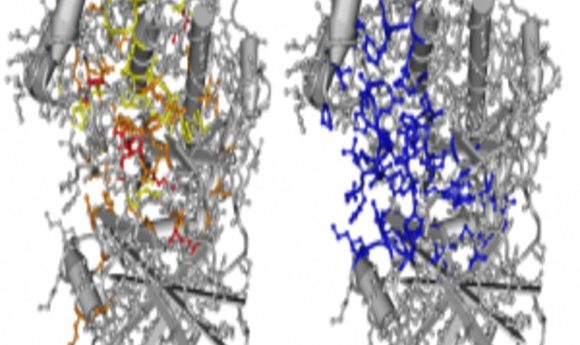
Oncodomain hotspots overlap significantly with protein active sites.
Credit: Thomas A. Peterson
Genetic mutations in cells can prompt abnormal growth, leading to tumor formation. Although some cancer-targeting drugs attack the associated mutated proteins, scientists so far have identified only a tiny portion of the rare mutations that contribute to cancer. Now, writing in PLoS Computational Biology, translational bioinformatics researcher Maricel Kann at the University of Maryland proposes a more efficient statistical analysis method for pinpointing rare cancer-causing mutations on a broader scale (1).
“Our prior study showed [that] you can define hotspots at the protein domain level from mutations involved in both Mendelian diseases and cancer, particularly for oncogenes,” said Kann (2). “Here, we addressed the next logical question: What happens when we don’t know the relevance of the mutations for cancer?”
Instead of focusing on individual gene mutations as earlier studies have, Kann’s method tackles related mutations in common domains across families of proteins. After mapping somatic variants from over 5,800 patients with 20 cancer types, her team proposed the term “oncodomain” to encapsulate the hotspots they found.
“Single gene mutations have limited usefulness because many are uncommonly found,” she explained. “Since domains are the functional units of a protein, functionally based approaches like ours might be the only way to detect these rare variants. Of course, if done improperly, this could lead to false positives. But like all analyses, the devil is in the details.”
Using this method, Kann’s team realized that the mutations occurred in the same domain location as mutations from other tumor-associated proteins, suggesting their involvement in cancer-linked pathways like protein phosphorylation, gene regulation, and cell metabolism.
“We were surprised by how many more variants and genes we detected with this approach,” Kann noted—over 5000 genes, 3000 of which are new finds but share structural and functional similarities with known cancer genes.
“Nature doesn’t reinvent the wheel each time it needs a function; instead, it re-uses what’s been created previously in other species or even within the same organism,” explained Kann. Because domains look similar across so many proteins, additional explorations of these oncodomains could direct existing research to characterize cancer types and improve broad-spectrum cancer drug development.