Researchers begin to explain counterintuitive spindle movement
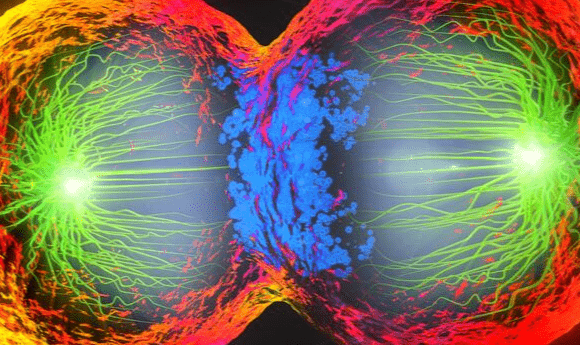
A new theory could explain how spindles have defied previous attempts at modeling their movement.
A researcher collaboration including the Flatiron Institute’s Center for Computational Biology (CCB; NY, USA), Harvard University, MIT (both MA, USA), Indiana University (IN, USA) and the University of California, Santa Barbara (CA, USA) may have provided an explanation for the discrepancy between the theorized and observed behavior of the molecular machinery that governs chromosomal splitting during cell division. The research, published in Nature Physics, proposes that the observed congestion of spindle fibers is what allows the spindle clusters to continue to move at full speed as opposed to the expected crawl.
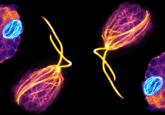
Spindles are responsible for dividing the chromosome during cell division, ensuring that both daughter cells contain a full set of genetic material. The main components of spindles are microtubules cross linked with kinesin molecular motors. Errors in the formation and movement of this cellular machinery can introduce errors into the cell such as extra or missing chromosomes which can lead to future complications such as infertility or cancer.
The properties of microtubules have been extensively characterized. They have a plus and a minus end and are mobilized by kinesin molecular motors. Kinesins have two pairs of feet, one at each end, that can attach two microtubules to each kinesin motor simultaneously. When the plus and minus end of two microtubules are aligned, the microtubules don’t move relative to one another as the movement of the kinesins is polar. When the microtubules are anti-aligned, the feet move in opposite directions and therefore slide past each other. As a result, they are only propelled when connected to a neighboring microtubule pointing in the opposite direction. However, previous research identified that even when linked to neighbors orientated in the same direction, they continued to move at full speed.
- A tale of two spindles
- Centromeres: the final frontier
- Modeling mitosis: how combining proteins could help in understanding dividing cells
Previous research assumed that the kinesin motors were relatively scarce within the cell and that microtubule movement depended on the orientation of their neighbor. As a result, it was assumed that the aligned microtubules should stay put while those that weren’t should remain stationary. Observations have highlighted that real spindles do not exhibit this behavior.
“It’s like a New York City crosswalk,” commented Sebastian Fürthauer (CCB). “People walking different ways are all mixed together, yet everyone is able to move at full speed and flow smoothly past one another.”
The researchers investigated how microtubules would move if the system was packed with motors. They developed a mathematical theory of how mechanical stresses develop with the collective when microtubules are pushed and pulled by multiple motors.
The theory predicts that the microtubules line up with everyone facing one of two opposite directions. Microtubules with opposite orientation mingle and as expected are propelled forwards. The theory states that microtubules elsewhere in the bundle are so entangled with their neighbors that they are pulled along by the progressing tubules. This results in every microtubule moving at the precise walking speed of the microtubule motor regardless of their place in the crowd.
The researchers experimentally validated this theory by using microtubules and abundant kinesin motors. They discovered that the predictions and experiments matched real-world spindles, most notably in the eggs of African clawed frogs. The spindles were observed to move at roughly the same speed that the motors connecting the microtubules are known to move at.
The researchers hope that this research can provide the framework for future research into spindle formation and movement. It could allow them to begin work on constructing their own spindles or more accurately construct computer models of these intricate cellular machines.