Paramagnetic NMR in structural biology

In this editorial, Enrico Ravera gives an overview of the use of paramagnetic metals in NMR and explains how this can provide crucial information about the structure and dynamics of proteins, nucleic acids and their complexes.

Dr Enrico Ravera joined the faculty at the Department of Chemistry of the University of Florence (Italy)as an assistant professor in June 2017. He graduated with a BSc in Chemistry from the University of Florence in 2008 under the guidance of Professor Roberta Pierattelli and Professor Ivano Bertini. He graduated with a MSc in Chemistry of Biological Molecules cum laude in 2009 and a PhD in Chemistry (General and Inorganic) in 2013 from the same University, under the guidance of both Professor Claudio Luchinat and Professor Ivano Bertini.
He was a Postdoctoral fellow from 2013–2015 at the Magnetic Resonance Center (CERM) at the University of Florence and in 2016–2017 was the recipient of a FIRC Fellowship at CIRMMP (Florence, Italy).
His research interests include NMR in solution and in solids – with particular reference to paramagnetic systems and biomaterials, EPR spectroscopy, and integrated methods for structure and dynamics of biomolecules. He is co-author of the book “NMR of Paramagnetic Molecules” (Elsevier, 2016) with I. Bertini, C. Luchinat and G. Parigi and co-editor of the book “Paramagnetism in Experimental Biomolecular NMR” (RSC Publishing, 2018) with C. Luchinat and G. Parigi.
NMR spectroscopy is an irreplaceable tool for structural biology because it can provide access to information about both the structure and the dynamics of biomolecular systems (1). However, solving a NMR structure from scratch is not a particularly easy task, because it requires extensive resonance assignment to obtain meaningful distance restraints.
In this context, paramagnetism-based restraints offer high-quality data that may be obtained with minor experimental efforts on many biologically-relevant systems as a large proportion of proteins are metalloproteins.
Among them, several inherently contain paramagnetic metals (such as copper(II) or iron(III)) (2), and for several other a native diamagnetic metal can be substituted by a paramagnetic one (e.g. cobalt(II) for zinc(II), lanthanoids(III) for calcium(II), and so on) (3, 4). Many groups worldwide have put in much effort towards the development of metal-binding tags that can be covalently bound to proteins and nucleic acids (5–7).
The presence of paramagnetic species imparts changes on the appearance of the NMR spectrum, which range from very small to dramatically large. These effects are a) contact shifts (CS), b) pseudocontact shifts (PCS), c) residual dipolar couplings (RDC) and d) paramagnetic relaxation enhancements (PRE) (8).
CS can observed for nuclei within a few bonds from the metal center and experience some unpaired electron spin density (9). They are usually rather large and associated to short (1–10 ms) nuclear relaxation rates. They provide information on i) the electronic structure at the metal center (hence the coordination geometry) and ii) the relative position of those nuclei with respect to the metal center. CS have, for instance, proven useful to understand the charge distribution in iron–sulfur clusters, hence indicating the mechanism for electron transfer or cluster assembly processes (10–12). CS can be observed in solid-state NMR as well and it is so sensitive to minor rearrangements that it was possible to observe the doubling of the shifted resonances in a case where the protein crystallized with two molecules in the asymmetric unit of the crystal (13).
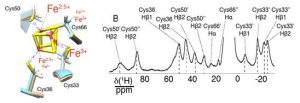
Figure 1. Superposition of the Fe4S4 cluster and the coordinating cysteines in the two molecules present in the asymmetric unit of EhHiPIP I and 1H double-adiabatic echo spectrum. (‘) and (“) denote the two different forms resulting from resonance doubling, each corresponding to one of the two molecules in the asymmetric unit. Adapted with permission from (13). Copyright 2017 American Chemical Society.
PCS result from the isotropic average of the additional field that is generated by the presence of the paramagnetic center, therefore they are related to the anisotropy of the magnetic susceptibility of the metal center (14, 15). They have an r-3 dependence on the metal–nucleus distance and also depend on the position of the nucleus in the frame defined by the magnetic susceptibility tensor. PCS can be observed in the solid state as well (16, 17).
Paramagnetic molecules are also attracted by the applied field and if their magnetic susceptibility is anisotropic, it is relatively easy to imagine that different orientations of the molecule with respect to the magnetic field will have slightly different energies. In solution NMR, this results in a partial alignment of the molecule in the field, which in turn results in non-zero averaging of the nuclear dipole–dipole interactions (RDC) (18).
PCS and RDC are long-range in nature and therefore are among the most popular restraints to achieve the structural determination of proteins, nucleic acids, and their complexes (19). Protocols for their use have been introduced in the most common structural determination and refinement software such as Cyana, (20) X-plor NIH (21) and REFMAC (22).
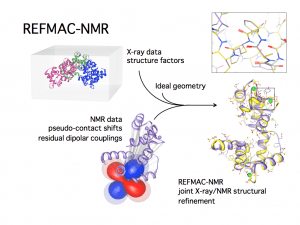
Figure 2. Scheme of the REFMAC-NMR joint refinement of a structure with X-ray reflections and NMR data: the local information provided by the NMR data changes in a non-trivial way the coordinates of the heavy atoms, yielding a more accurate structure (figure courtesy of Dr. Azzurra Carlon).
The presence of a paramagnetic center also increases the relaxation rates of nuclei close to the unpaired electron spin(s), depending on the electron relaxation properties and thus on the electronic structure, because a further mechanism for nuclear relaxation becomes operative, i.e. the modulation of the dipole–dipole interaction between electron and nuclear spins (23–25). This increase in the nuclear relaxation rates is the PRE, which is proportional to the sixth power of the distance from the metal center. Because of this very steep dependence, PRE are very effective in detecting transient, lowly populated states (26, 27). For instance, PREs were instrumental to identify plausible minor states, which are populated for around 1% of the time collectively, in the electron transfer encounter complex between putidaredoxin and cytochrome P450cam (28). Interestingly, some of the minor states seem to be connected to the main state through electrostatic runways (28, 29).
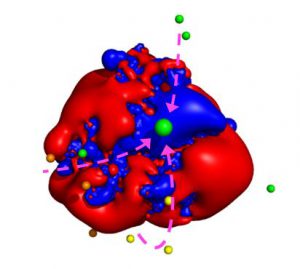
Figure 3. Plausible center of mass positions of putidaredoxin in the minor states as identified by the use of PRE (indicated by the smaller spheres) with respect to the positive (blue) and negative (red) electrostatic potential isosurfaces of cytochrome P450cam. The larger green sphere represents the main state. The arrows indicate possible paths connecting some of the minor states to the main state (productive encounters) whereas others remain isolated (futile encounters). Reproduced from Journal of Magnetic Resonance, 282, Enrico Ravera, Giacomo Parigi, Claudio Luchinat, Perspectives on paramagnetic NMR from a life sciences infrastructure, 154, Copyright (2017), with permission from Elsevier.
Paramagnetic data are an important source of structural restraints in integrated structural biology, because they are essentially unambiguous: they can be easily obtained provided that the assignment of each single signal of the analog diamagnetic protein is available. Since PCS arising from metals with large magnetic susceptibility anisotropy are experimentally measurable even at relatively large distances from the metal, as well as PRE due to metals with large electron relaxation time (despite the r-6 dependence), they are very useful to calculate a preliminary structural model of the molecule, possibly complemented by chemical shift information, which permits us to resolve ambiguities in the NMR spectra, so that other (paramagnetic and diamagnetic) restraints can be obtained.
In conclusion, all these compelling features are pushing more and more structural biologists towards the use of paramagnetic restraints, rapidly promoting them from a specialist’s toolbox to the common use in everyday research. NMR is extremely sensitive to local rearrangements, thus it is extremely complementary to X-ray diffraction. By the same token, it is clear that NMR is complementary to cryo-electron microscopy as well. Therefore it is to be expected that, in the near future, we will witness and participate in the blooming of integrative approaches.
Sources:
- Bertini I, McGreevy KS, Parigi G. NMR in systems biology (Wiley) (2012)
- Bertini I, Gray HB, Stiefel EI, Valentine JS. Biological Inorganic Chemistry: Structure and Reactivity. (University Science Books) (2006)
- Bertini I, Luchinat C. High spin cobalt(II) as a probe for the investigation of metalloproteins. Adv. Inorg. Biochem. 6:71–111 (1985)
- Allegrozzi M, Bertini I, Janik MBL, Lee Y-M, Liu G, Luchinat C. Lanthanide induced pseudocontact shifts for solution structure refinements of macromolecules in shells up to 40 A from the metal ion. J. Am. Chem. Soc. 122:4154–4161 (2000)
- Wöhnert J, Franz KJ, Nitz M, Imperiali B, Schwalbe H. Protein Alignment by a Coexpressed Lanthanide-Binding Tag for the Measurement of Residual Dipolar Couplings. J. Am. Chem. Soc. 125:13338–13339 (2003)
- Pintacuda G, John M, Su X-C, Otting G. NMR Structure Determination of Protein−Ligand Complexes by Lanthanide Labeling. Acc. Chem. Res. 40:206–212 (2007)
- Keizers PHJ, Ubbink M. Paramagnetic tagging for protein structure and dynamics analysis. Prog. Nucl. Magn. Reson. 58:88–96 (2011)
- Bertini I, Luchinat C, Parigi G, Ravera E. Solution NMR of paramagnetic molecules: applications to metallobiomolecules and models (2017)
- McConnell HM, Chesnut DB. Theory of Isotropic Hyperfine Interactions in π‐Electron Radicals. J. Chem. Phys. 28:107–117 (1958)
- Banci LI, Bertini LD, Eltis IC Felli, D.H.W. Kastrau, C. Luchinat, M. Piccioli, R. Pierattelli, and M. Smith. The three dimensional structure in solution of the paramagnetic protein high-potential iron-sulfur protein I from _Ectothiorhodospira halophila_ through nuclear magnetic resonance. Eur. J. Biochem. 225:715–725 (1994)
- Banci L, Bertini I, Ciurli S, Luchinat C, Pierattelli R. Rationalization of the reduction potentials within the series of the high potential iron-sulfur proteins. _Dedicated to Prof. F.Basolo_. Inorganica Chim. Acta. 240:251–256 (1995)
- Brancaccio D, Gallo A, Mikolajczyk M et al. Formation of [4Fe-4S] Clusters in the Mitochondrial Iron–Sulfur Cluster Assembly Machinery. J. Am. Chem. Soc. 136:16240–16250 (2014)
- Bertarello A, Schubeis T, Fuccio C et al. Paramagnetic Properties of a Crystalline Iron-Sulfur Protein by Magic-Angle Spinning NMR Spectroscopy. Inorg. Chem. 56:6624–6629 (2017)
- McConnell HM, Robertson RE. Isotropic Nuclear Resonance Shifts. J. Chem. Phys. 29:1361–1365 (1958).
- Kurland RJ, McGarvey BR. Isotropic NMR shifts in transition metal complexes: calculation of the Fermi contact and pseudocontact terms. J.Magn.Reson. 2:286–301 (1970)
- Balayssac S, Bertini I, Lelli M, Luchinat C, Maletta M. Paramagnetic Ions Provide Structural Restraints in Solid-State NMR of Proteins. J. Am. Chem. Soc. 129:2218–2219 (2007)
- Bertin I, Emsley L, Lelli M, Luchinat C, Mao J, Pintacuda G. Ultrafast MAS Solid-State NMR Permits Extensive 13C and 1H Detection in Paramagnetic Metalloproteins. J. Am. Chem. Soc. 132:5558–5559 (2010)
- Lohman JAB, Maclean C. Alignment effects on high resolution NMR spectra induced by the magnetic field. Chem. Phys. 35:269–274 (1978)
- Bertini I, Luchinat C, Parigi G, Pierattelli R. Perspectives in NMR of paramagnetic proteins. Dalton Trans. 2008:3782–3790 (2008)
- Banci L, Bertini I, Cremonini MA et al. PSEUDYANA for NMR structure calculation of paramagnetic metalloproteins using torsion angle molecular dynamics. J. Biomol. NMR. 12:553–557 (1998)
- Banci L, Bertini I, Cavallaro G, Giachetti A, Luchinat C, Parigi G. Paramagnetism-based restraints for Xplor-NIH. J. Biomol. NMR. 28:249–261 (2004)
- Rinaldelli M, Ravera E, Calderone V, Parigi G, Murshudov GN, Luchinat C. Simultaneous use of solution NMR and X-ray data in REFMAC5 for joint refinement/detection of structural differences. Acta Crystallogr. D Biol. Crystallogr. 70:958–967 (2014)
- Solomon I. Relaxation Processes in a System of Two Spins. Phys. Rev. 99:559–565 (1955)
- Gueron M. Nuclear relaxation in macromolecules by paramagnetic ions: a novel mechanism. J.Magn.Reson. 19:58–66 (1975)
- Vega AJ, Fiat D. Nuclear relaxation processes of paramagnetic complexes The slow-motion case. Mol. Phys. 31:347–355 (1976)
- Clore GM, Iwahara J. Theory, Practice, and Applications of Paramagnetic Relaxation Enhancement for the Characterization of Transient Low-Population States of Biological Macromolecules and Their Complexes. Chem. Rev. 109:4108–4139 (2009)
- Bashir Q, Volkov AN, Ullmann GM, Ubbink M. Visualization of the Encounter Ensemble of the Transient Electron Transfer Complex of Cytochrome c and Cytochrome c Peroxidase. J. Am. Chem. Soc. 132:241–247 (2010).
- Andrałojć W, Hiruma Y, Liu WM et al. Identification of productive and futile encounters in an electron transfer protein complex. Proc. Natl. Acad. Sci. U. S. A. (2017)
- Ravera E, Parigi G, Luchinat C. Perspectives on paramagnetic NMR from a life sciences infrastructure. J. Magn. Reson. 282:154–169 (2017)