A peek behind the paper – David Saul and Jo Stanton on the new method for DNA extraction

Take a look behind the scenes of a recent Reports article, entitled: ‘Rapid extraction of DNA suitable for NGS workflows from bacterial cultures using the PDQeX’, as we ask authors about PDQeX, a new method for DNA extraction.

Senior Research Fellow, Jo Stanton (University of Otago; New Zealand), uses nucleic acid-based technologies to explore a range of scientific interests related to infectious disease, reproduction, the microbiome, anthropology and forensics. She leads a team of cross-disciplinary scientists from both academia and industry to develop rapid, accurate and cost-effective handheld molecular diagnostic devices and systems for in-field and point-of-care applications.
Jo is one of New Zealand’s experts in high-throughput sequencing technologies, having established the first fully-operational New Zealand HTS Services based around the 454 Roche GS FLX. This service expanded to include the SOLiD platform from Life Technologies (CA, USA). She currently works with MinION technology from Oxford Nanopore (UK) and MicroGEM’s (UK) PDQeX Nucleic Acid Extractor to adapt their use for point-of-care and in-field situations.
Jo received a BSc (Honors) from the Australian National University (ACT, Australia) and a PhD from the University of Western Australia (WA, Australia).

Chief Scientific Officer, David Saul’s (MicroGEM) expertise spans molecular genetics, microbiology, biotechnology, computational biology and forensic DNA analysis. His key research interest has been the development of biotechnical applications of microorganisms living in extreme environments. With a focus on DNA detection and identification, David has generated a substantial body of intellectual property around processes and devices for simple, low-cost and competitive diagnostics.
Prior to co-founding ZyGEM (now MicroGEM), David worked as a researcher and academic at the University of Auckland (New Zealand). He has worked in several fields relating to the molecular study of organisms and their genes. He graduated with a PhD in molecular genetics from The University of Sheffield (UK).
What inspired you to write this piece?
Data quality from all nucleic acid tests rests squarely with the quality of the input DNA. Up until now, methods for nucleic acid extraction have either required the use of toxic chemicals or have taken several hours to perform. There has also been sample loss due to multiple pipetting steps, centrifugation, etc. We, therefore, wanted to drive toward a single step nucleic acid purification system that was quick, reliable and gave consistent output.
In addition, the read-length of extracted DNA has become more important. With Illumina sequencing, sheared DNA is less of a problem. But library insert sizes are usually around 1200 base pairs of which 150 bases are read from each end. The improving reliability and cost of nanopore sequencing (MinION) and SMRT sequencing (PacBio) means that more intact DNA is needed, so sample preparation that results in less shearing is important.
Coupled with these needs, current technologies tie nucleic acid extraction to the laboratory yet the paradigm shift of moving nucleic acid-based tests closer to the point-of-need is transformative. Think of border control officers being able to screen for CITES protected species at their point of entry or farmers needing fast answers about plant pathogens or crop-damaging insects. Reliable nucleic acid testing at the point-of-need starts with quick, robust, and reliable sample preparation systems designed for use in resource-limited settings.
What is PDQeX?
The PDQeX is a revolutionary new way to extract DNA from a sample. It consists of three component parts: (i) a cocktail of enzymes isolated from mesophilic and extremophilic organisms that use temperature to control the reaction; (ii) an innovative extraction cartridge consisting of an inner and outer tube and a filter unit (the inner tube is made of thermo-responsive plastic that shrinks at high temperature, forcing the extract through the filter); and (iii) a thermal control unit named the PDQeX Nucleic Acid Extractor.
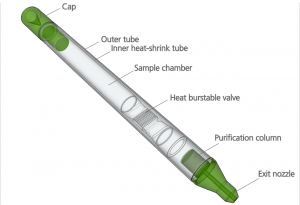
How does your new method differ from current DNA extraction methods?
Handling is much simpler than current DNA extraction methods because the extraction steps are carried out in a single tube by the biochemistry and temperature shifts. And because the chemistry is compatible with most reaction buffers, it means you don’t need all the purification steps. It’s quicker – sample preparation takes minutes, not hours – and gentler so there’s less shearing. Current methods often require harsh and toxic chemicals, while multiple pipetting and centrifugation steps result in several hours of sample preparation.
In the PDQeX system, the sample is simply added to the extraction tube along with the enzyme cocktail. The PDQeX extraction tube is capped with a lid and placed into the thermal control unit. The assembly is heated in increasing temperature steps that activate the different extremophile enzymes. These enzymes sequentially degrade the cell wall and lyse the tissue (35 or 52°C) then protein is digested (75°C). Another rise in temperature causes the tube to shrink, pushing the lysate through a purification polymer that filters the DNA and removes inhibitors, like polyphenolics and carbohydrates, and the enzymes used for extraction. The whole process takes less than 20 minutes with no hands-on operational steps.
Webinar: Rapid single-tube preparation of DNA from bacteria for NGS workflows
What impact do you see your new method having on genome sequencing?
The PDQeX will impact all aspects of genome sequencing. One good example is using genome sequencing as a point-of-need diagnostic tool.
We have coupled the PDQeX with sequencing technology from Oxford Nanopore to diagnose cassava mosaic virus on remote farms in East Africa. Sequencing requires good quality DNA, and DNA isolation has, up until now, been a stumbling block for in-field sequencing. Other researchers have simply replicated standard DNA extraction methods in a tent, requiring considerable infrastructure and power requirements. The PDQeX only requires a battery to extract DNA. The enzymes are lyophilized removing any requirement for a cold chain. Extracted DNA can be used directly with the Oxford Nanopore technology. In our study, virus detection, including viral type information, was diagnosed within 4 hours of arrival on the farm. This demonstration has broad implications, for example, in agriculture, biosecurity, food safety, and curbing the illegal trade in plants and animals. This enables genome sequencing to be removed from the remote laboratory and performed on site, where and when information is needed.
What are you hoping to focus your research on over the next 5 years?
Our team is developing fully integrated, portable instruments to provide answers at the point-of-care or in the field – with minimal training.
Tell us a little about the company behind this technology.
DNA and RNA hold the keys to resolving many of the toughest research challenges. MicroGEM’s approach to faster, nearly effortless, nucleic acid extractions empowers researchers to move quickly to their important work of treating and curing diseases; addressing water quality, crop health and food security; tackling crime scene analysis and human ID; and advancing many of the health, safety and environmental issues we face as a society.
MicroGEM helps scientists move quickly to analysis, produce faster results, and make timely decisions about the implications of their studies. Our temperature-driven approach sets the standard for DNA and RNA extractions where speed, sample size, yield, and simplified workflow are important. Single-tube extractions produce high-quality nucleic acids in minutes, not hours, without the overuse of plastic or harsh chemicals. Reduced handling means fewer opportunities for contamination, all at an affordable cost.
In addition to PDQeX, MicroGEM offers a full range of extraction kits suitable for use with existing laboratory equipment and many different sample types including cell culture, tissue, bacteria, insects, and sperm. The enzyme at the heart of this chemistry is prepGEM™, a reagent used extensively for DNA extractions. It has become the gold standard for the extraction of nucleic acids for library preparation in NGS since reaction volumes can be kept small and there is no loss of DNA.
Find more information about PDQeX and temperature-driven enzymatic extractions at https://microgembio.com/
You can read the full article on the journal website here: Rapid extraction of DNA suitable for NGS workflows from bacterial cultures using the PDQeX And can learn more about the method at our upcoming webinar: Rapid single-tube preparation of DNA from bacteria for NGS workflows
Reusing this method? Follow the Reuse Recipe here: https://bit.ly/2Y9kVDD