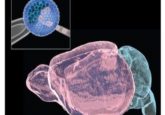
Targeting tomato shoot regeneration

MicroRNA396 has been identified as a negative regulator of shoot regeneration in tomatoes, unlocking possibilities for plant genetic engineering.
Researchers from the Korea Research Institute of Bioscience and Biotechnology (Daejeon, South Korea) last year identified microRNA396 (miR396) as a negative regulator of shoot regeneration in tomatoes. Targeting this single RNA could improve tomato shoot regeneration efficiency and help create gene-edited plants in genotypes with low regeneration efficiency.
Shoot regeneration is a central component of genetic engineering, with many plants generated by inducing shoot regeneration through the reprogramming of transformed somatic cells. However, shoot regeneration efficiency is highly dependent on genotypes, and the molecular mechanisms behind this are still not fully understood.
Key genes involved in shoot regeneration in various crop species have previously been identified, including the growth regulating factor (GRF) family genes. While genetically engineering plants to overexpress a specific GRF can improve plant regeneration, selecting the effective GRF gene requires individual studies for each plant species. Therefore, uncovering widely conserved upstream regulators of GRFs that determine high- and low-efficiency regeneration genotypes could provide a target for genetic engineering of tomatoes and other crops.
 Sowing the seeds of sustainable agriculture and medicinal plant research
Sowing the seeds of sustainable agriculture and medicinal plant research
Sylvia Mitchell, Head of the Medicinal Plant Biotechnology Research Group at the University of the West Indies, shares her experiences working in medicinal plant research, highlighting the techniques she uses for plant tissue culturing and biochemical analyses, the importance of harnessing native plant species for sustainable practices and the commercial aspects of her field of research.
To investigate this, the researchers selected two tomato genotypes, Sweet King (SK) and Super Doterang (SD), which show highly different regeneration capacities. The low-efficiency SK genotype has a regeneration capacity of 14% after 42 days of incubation, while the high-efficiency SD genotype has a regeneration capacity of 97%. Time-course transcriptome analysis of the two genotypes was performed to analyze the differentially expressed genes, followed by small RNA-sequencing of differentially abundant miRNAs to find the upstream regulators of these genes. Comparative analysis of the data revealed miR396 to be a key molecule in determining shoot regeneration efficiency.
MicroRNA396 and its target transcripts encoding GRFs showed differences in abundance between high- and low-efficiency genotypes, with GRFs elevating 12–18 days after incubation in SD but remaining largely unaltered in SK. Conversely, the abundance of miR396 in SK was significantly higher than in SD during incubation.
The researchers then suppressed miR396 in tomato explants and observed improved shoot regeneration efficiency. Further, they discovered that gene-edited plants could be generated in the low-efficiency SK tomato genotype by co-transforming vectors expressing Cas9-gRNA and STTM396, a miR396 suppressor.
“Our findings reveal the critical role of miR396 in tomato shoot regeneration and suggest that by blocking this single microRNA, we can significantly improve regeneration efficiency. This opens new possibilities for genetic engineering in tomatoes and potentially other crops,” commented corresponding author Hyun-Soon Kim.
These results have helped to increase understanding of the molecular mechanisms that occur during tomato shoot regeneration and highlight future possibilities of miR396 in plant genetic engineering.