Proteins take the heat
A new study shows how cranking up the heat causes cell death through the loss of a subset of key proteins.
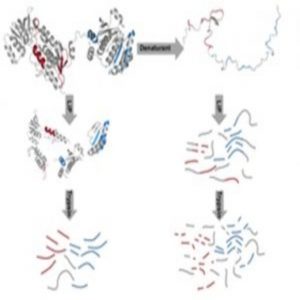
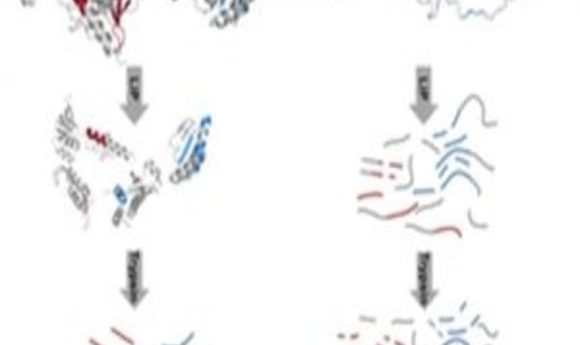
Workflow of the LiP-MS-based analysis of protein stability (1).

Thermophiles thrive at temperatures up to 122oC.
Cells are picky with regards to temperature. While human cells are cozy at 37°C, so-called thermophiles love the heat and thrive at temperatures between 41°C and 112°C. Beyond basic cell function, thermostability also has clinical and industrial applications. Heat damages cancer cells and kills bacteria in foods; microorganisms genetically engineered to take the heat are used in laundry detergents and bioethanol production.
“What makes an organism collapse upon an increase in temperature, and why do certain species survive even at high temperatures?,” wondered Paola Picotti from ETH Zurich, senior author of a new study published in Science.
To study the culprits in cell death, the researchers turned to proteins—the most abundant and least stable molecules in a cell. Their goal was to measure the thermostability of a multitude of proteins simultaneously across different cell types, but to do so required a novel method.
First, the researchers exposed three cell types, Escherichia coli,the heat-loving bacterium Thermus thermophiles, and human-derived HeLa cells, to a range of temperatures known to denature proteins. Next, the researchers collected the cells and performed limited proteolysis (LiP) on the proteome extracts by applying a non-specific protease to the mix of cells and then prematurely stopping the digestion, generating structure-specific protein fragments. “If a protein changes structure upon a perturbation, you will see a completely different proteolytic pattern,” said Picotti. “So we reasoned that if a protein undergoes progressive unfolding, for example due to a temperature increase, then we should see more and more proteolytic cleavage.” The researchers measured proteolytic cleavage with mass spectrometry (MS), enabling them to reconstruct the thermal unfolding of a protein.
At temperatures that impair cell function, only a subset of essential proteins undergo denaturation, contradicting the researchers’ prediction that proteome instability results from the loss of hundreds of proteins.
Further analysis of the data revealed that the most thermodynamically stable proteins are also the most abundant. These findings provide experimental evidence for the “translational robustness” theory, which posits that highly expressed proteins must be able to tolerate the number of translational errors that can occur in a cell.
Picotti and colleagues plan to further explore this limited population of unstable proteins more closely. “It would be interesting to try to genetically modify these proteins to make them more thermostable and then see if you can obtain an organism that can live at higher temperatures than its natural living condition,” said Picotti.