Identifying CRISPR-generated mutants using high-resolution melting analysis

Genome modification has been greatly facilitated by the CRISPR/Cas9 system, which has simplified targeted mutagenesis by significantly increasing efficiency and specificity. A critical step in any CRISPR/Cas9 mutagenesis workflow is screening for induced mutations.
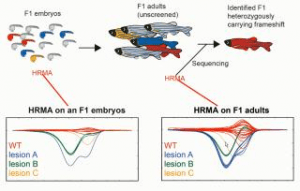
In this tutorial webinar, Jared C. Talbot, a researcher at The Ohio State University, describes how high-resolution melting analysis (HRMA) can be used to screen for insertion/deletion (indel) mutations generated by CRISPR/Cas9.
Specifically, you will learn:
- How to design a CRISPR/Cas9 pipeline for creating indel mutations
- The basics of using HRMA
- Tips and tricks for using HRMA to identify CRISPR-generated indel mutations

Featured speaker:
Jared Talbot
The Ohio State University
Dr. Talbot is a post-doctoral fellow in the lab of Sharon Amacher at The Ohio State University, where he is currently studying the signaling processes that trigger muscle cell maturation in zebrafish.
He obtained his bachelor’s in biology from Cornell University, where he focused on development and genetics. He then pursued doctoral studies in Charles Kimmel’s lab at the University of Oregon, where he used zebrafish to study skeletal development.
Sponsored by:
