Navigating the brain using spatial transcriptomics in health, disease and development
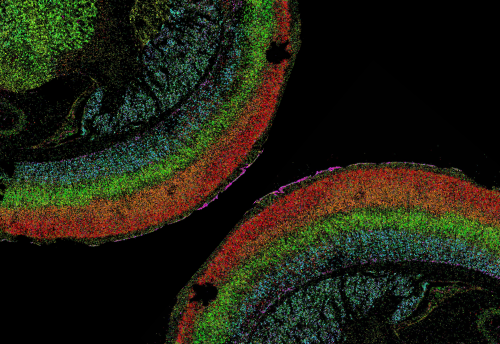
Elucidating cellular heterogeneity and the transcriptomic programs that shape neuronal diversity and circuit assembly is crucial to understanding the underlying neurobiology in health, disease and development.
Using Molecular CartographyTM, we quantified changes in transcriptomic activity and cellular phenotypes in response to Alzheimer’s disease pathology. This multiplexed, single-molecule detection technology also allows for the transcriptomic characterization of rare GABAergic interneurons in the developing mouse cortex with full spatial context at subcellular resolution.
What will you learn?
- How to perform highly multiplexed and sensitive spatial transcriptomics
- How to conduct spatial analysis of cell phenotypes in response to Alzheimer’s disease pathology
- How to characterize GABAergic interneurons in the developing brain
Who may this interest?
- Neuroscientists
- Developmental biologists
- Researchers who are interested in using spatial biology tools, single-cell technologies, in situ hybridization and imaging techniques to advance their research
- Anyone interested in simultaneously evaluating healthy and diseased samples.
Panelists

Jeroen Aerts
Customer Technology Advisor
Resolve Biosciences
Jeroen Aerts obtained his PhD in Neuroscience at KU Leuven (Belgium) in 2014, during which he specialized in histology, in situ hybridization and mass spectrometry imaging. After that, he did a Postdoc at Janssen Pharmaceutica (Beerse, Belgium), part of Johnson & Johnson, where he used single-cell sequencing in the context of drug development for Alzheimer’s disease.
In 2018, he joined VIB (Ghent, Belgium) Tech Watch where he scouted early-access technologies and tested beta-instruments for single-cell sequencing and spatial transcriptomics. During that time, he worked with many spatial technology companies and gained in-depth insights into the competitive spatial technology landscape. In July 2021, he joined Resolve Biosciences as a Customer Technology Advisor to help scientists advance their understanding of complex biological questions using theMolecular Cartography technology.

Lynette Lim
Group Leader & Assistant Professor of Neurosciences
Laboratory of Interneuron Developmental Dynamics
VIB Center for Brain & Disease Research
KU Leuven, Department of Neurosciences
Lynette Lim is a neurodevelopmental biologist, who works on how brain circuits are formed. Originally from Singapore, she obtained her PhD at the National University of Singapore in 2007, under Markus Wenk. Her thesis work focused on lipid metabolism in neurons.
In 2012, she was awarded a fellowship from the EMBO (Heidelberg, Germany) and joined the lab of Oscar Marin, first at the Insituto de Neurosciencia (Alicante, Spain) from 2012 to 2014, then at King’s College London (UK) from 2014 to 2019. During her postdoctoral training in the Marin lab, she published seminal findings on how early developmental programs shape brain wiring. Her work has been published in numerous top tier journals such as Science, Nature Neuroscience and Neuron, and she is a recognized emerging leader in the field for cortical circuit assembly. As such, in 2019, she was awarded, but respectfully declined, the Wellcome Trust Sir Henry Dale Fellowship.
In 2020, she was appointed group leader at VIB-CBD and Assistant Professor at KU Leuven. At VIB, her work has been successfully funded by the Flanders research foundation (FWO). In 2021, she was one of 13 top researchers who were awarded the flagship FWO Odysseus grant, which aims to encourage excellent researchers to come and work in Flanders and establish themselves within the Flemish research community. Her current research continues to focus on brain development, specifically on how different nerve cells emerge, mature and assemble into functional circuits in the mammalian neocortex.
This webinar was recorded on Thursday 7th Oct0ber 2021
In association with
